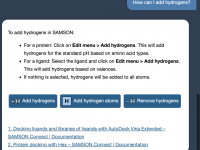
Author: OneAngstrom
Animating molecules
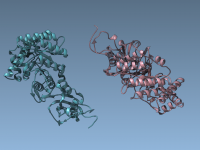
Generating symmetric replicas
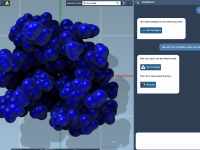
Making SAMSON AI ✨ do something
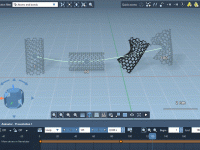
Creating an assembly animation
Editing side-chain conformations
Creating replicas of structures
Making molecules refractive
Quick geometry optimization using the FIRE minimizer
Accessing the data graph in Python
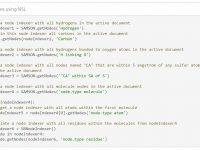
To efficiently manipulate nodes (atoms, bonds, side chains, molecules, etc.), use node indexers returned by the SAMSON.getNodes function applied to a Node Specification Language (NSL) expression. For example, SAMSON.getNodes(‘node.type atom’) returns an indexer containing all atoms in the active document.…