Why the FIRE Minimizer is a Game-Changer for Efficient Geometry Optimization

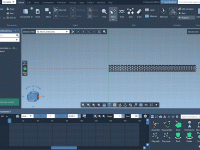
Geometry optimization is at the heart of molecular modeling, aiding scientists in achieving stable, accurate molecular structures for reliable simulations and designs. But it’s not uncommon for researchers to encounter slow convergence or inefficiencies when handling large-scale molecular systems with…
Smoothly Visualize Molecular Trajectories Using SAMSON’s Play Path Animation
Streamlining Molecular Design with Pattern Editors in SAMSON.
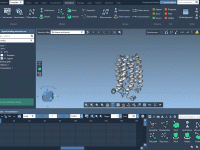
Molecular modelers often face the challenge of constructing complex nanoscale architectures from smaller units. Repetitive tasks like replicating and arranging molecular structures in specific configurations not only consume time but also increase the likelihood of errors. SAMSON’s Pattern Editors provide…
Efficiently Record and Export Atom Trajectories in SAMSON Animations
Molecular modelers often face the challenge of tracking and analyzing atomic trajectories during molecular animations. Whether you’re simulating molecular dynamics or presenting assembly processes, having a clear visualization of these trajectories can significantly enhance your understanding of molecular behavior. SAMSON…
Simplifying Molecular Presentations with the Hide Animation
Streamlining Ligand Unbinding Pathway Discovery with Sampling Regions
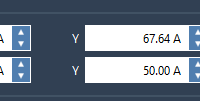
Mastering Molecule Attributes in SAMSON: Simplify Molecular Modeling
Refine Your Molecular Models: Exploring Molecule Attributes in SAMSON
Master Protein Symmetries with the Symmetry Mate Editor
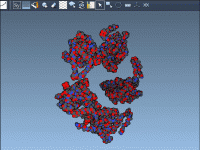
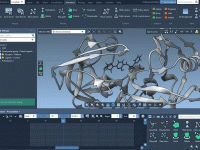
Analyzing protein-protein interactions and constructing realistic molecular models are everyday tasks for molecular modelers. However, reconstructing biological assemblies or understanding how proteins interact in their natural symmetric forms can be a daunting task without the right tools. The Symmetry Mate…