Generating reliable membrane-protein systems can quickly become a time-consuming task for molecular modelers. Between aligning proteins, setting up lipid orientations, tuning margins, and ensuring correct placement, a lot can go wrong. Fortunately, Molecular Box Builder in SAMSON simplifies the process of building lipid monolayers and bilayers around proteins in a controlled, visual environment.
This guide walks you through a specific and surprisingly smooth workflow: adding a lipid bilayer around a membrane protein structure, using just a few well-guided steps. The result? You can go from a plain PDB file to a spatially aware bilayer system and get ready for minimization and dynamics.
What You Need
- A protein structure (e.g., PDB: 4BBJ)
- A lipid molecule (POPC and others are available)
- SAMSON and the Molecular Box Builder Extension
Step 1: Align the Protein
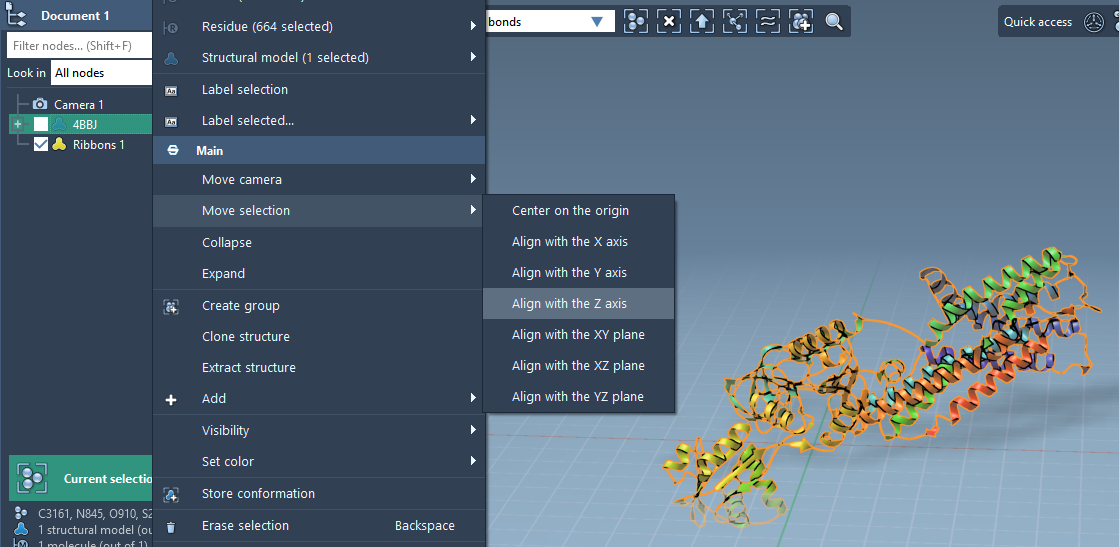
Alignment is essential to ensure proper membrane orientation. To reorient the protein in SAMSON:
- Right-click the protein in the Document view.
- Select Move selection > Align with Z axis.
- Then choose Move selection > Center on the origin.
Step 2: Set and Orient the Lipid
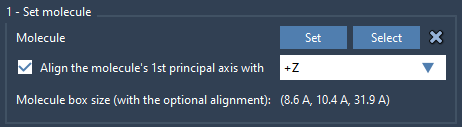
- Import a lipid molecule (you can use POPC or others).
- Select the lipid and open the Molecular Box Builder.
- Click Set to define it as the molecule to insert.
- Choose the +Z alignment for the lipid’s primary axis.
Step 3: Define the Lipid Box
- Center the box around the protein (you can match its centroid).
- Adjust the box height and width to fit a single layer of lipids, keeping an appropriate margin if needed.
- Enable the Consider existing molecules in the box option to avoid any lipid-protein overlap during insertion.
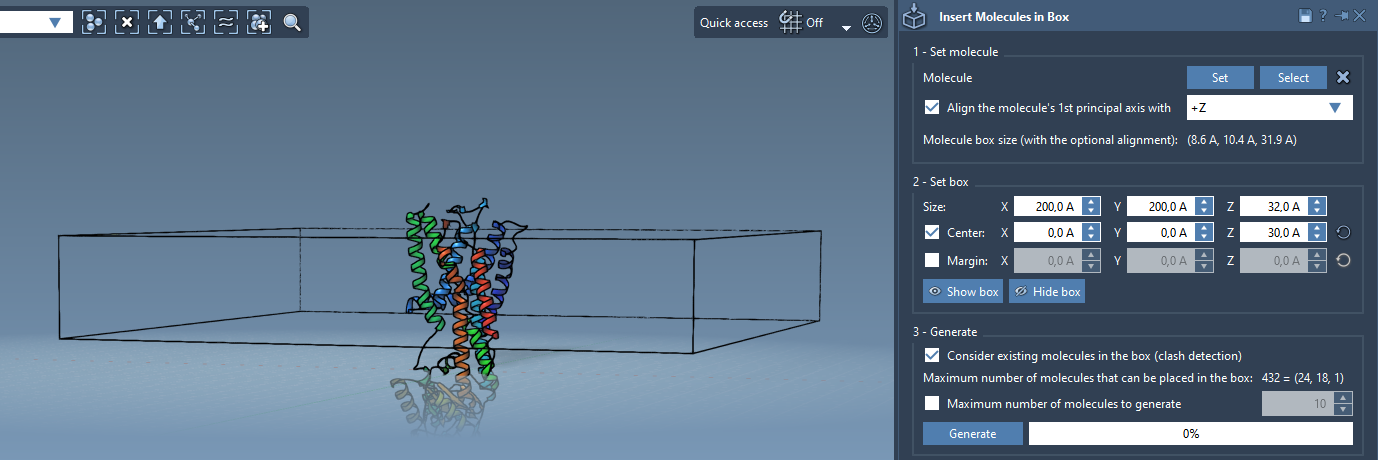
Step 4: Add the First Layer
Click Generate to add a single lipid layer on one side of your membrane protein:

Optional: Add the Second Layer
For a full bilayer:
- Create the first layer with +Z aligned lipids.
- Change the box center along the Z-axis (e.g., shift down).
- Switch the lipid alignment to -Z.
- Click Generate to place the second layer.
This yields a properly aligned bilayer membrane system, suitable for solvation or direct simulation preparation using tools like the GROMACS Wizard.
Why This Matters
This process saves hours of manual alignment and scripting, especially when building multiple models or screening membrane environments. The visual feedback from the Molecular Box Builder helps reduce placement errors and lets you explore different lipid types or protein orientations quickly.
To learn more or explore advanced options, visit the full Molecular Box Builder documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





