Master Protein Conformations with the Interactive Ramachandran Plot.
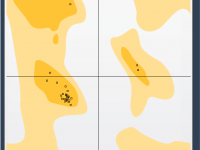
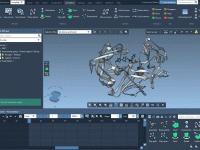
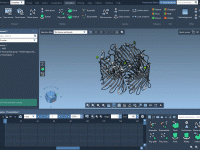
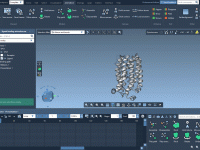
Validating and refining protein structures can be a challenging task for molecular modelers. Modeling accuracy directly impacts the success of simulations, predictions, and experimental designs. One particularly useful tool to address these challenges is the Interactive Ramachandran Plot provided by…