In medicinal chemistry and molecular modeling, exploring molecular variations through analog generation is often a key step in optimizing compounds. But modifying molecules atom-by-atom, especially when dealing with a high number of analogs, can be slow and error-prone if done manually. Fortunately, SAMSON offers an integrated solution: Positional Analogue Scanning using the SMILES Manager extension.
Let’s take a closer look at one important subset of this functionality: replacing or attaching atoms or groups at specific positions based on SMARTS patterns. This task plays a critical role in understanding how modifications alter molecular function—and this feature in SAMSON is designed to make it as accessible as possible.
When you know what to scan
Say you have a lead compound, and you’d like to systematically examine how replacing each aromatic hydrogen with a nitrogen or a methyl group affects biological activity. This is the essence of positional analogue scanning—and it’s now just a few clicks away in SAMSON.
How it works
Once your base molecule is loaded (via SMILES input or selection in the SAMSON document), you define a substructure pattern to search—typically, this is a SMARTS string. For example, [cH] captures aromatic carbons with a hydrogen, which are prime candidates for substitutions in SAR studies.

After selecting your pattern, you specify the replacement or attachment group: a nitrogen atom, fluorine, or a methyl group, for instance. Click Run, and the SMILES Manager generates a list of analogs with the specified modifications. Each variation is displayed with its 2D structure, simplifying visual assessment at the earliest stage.
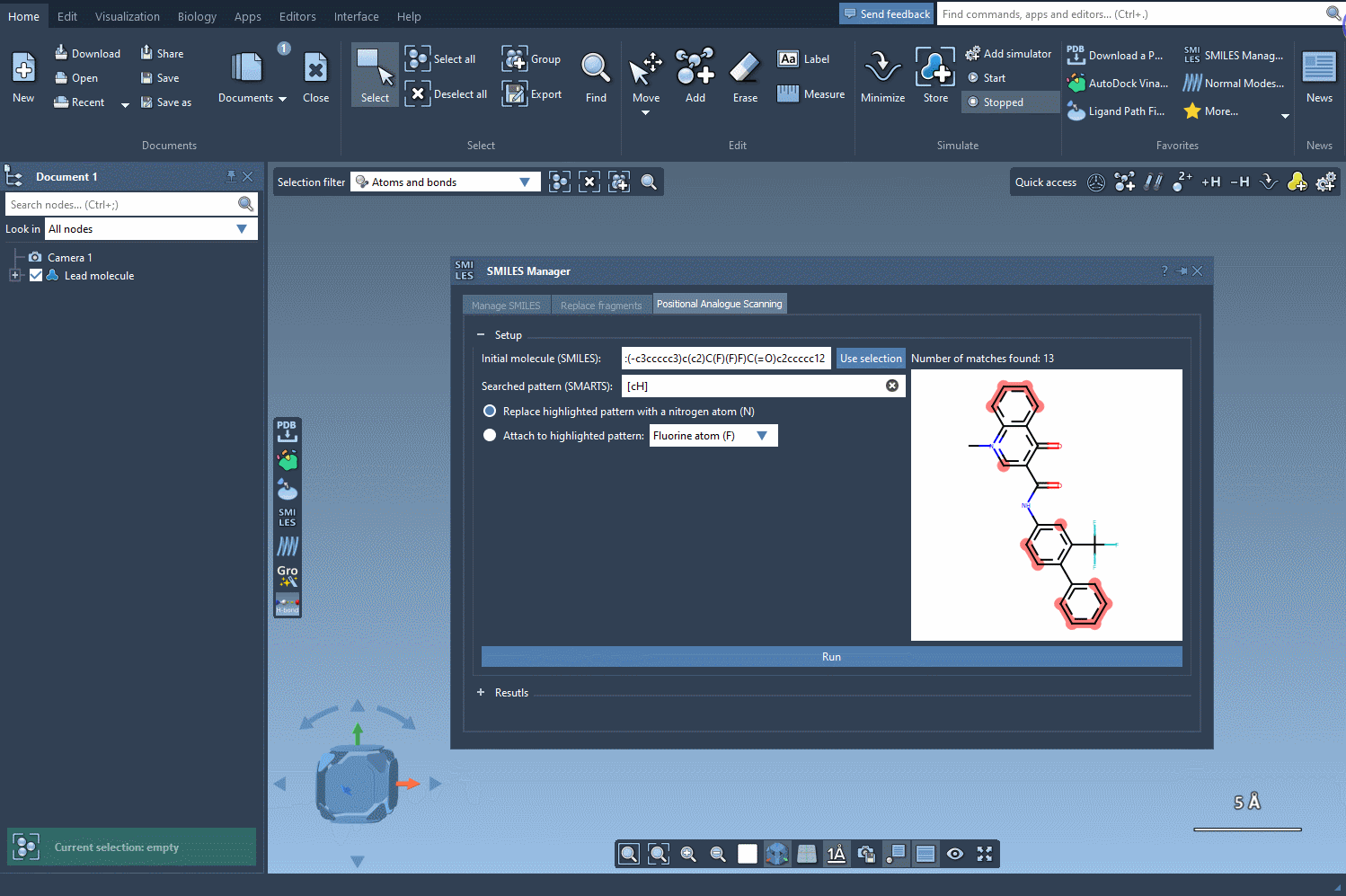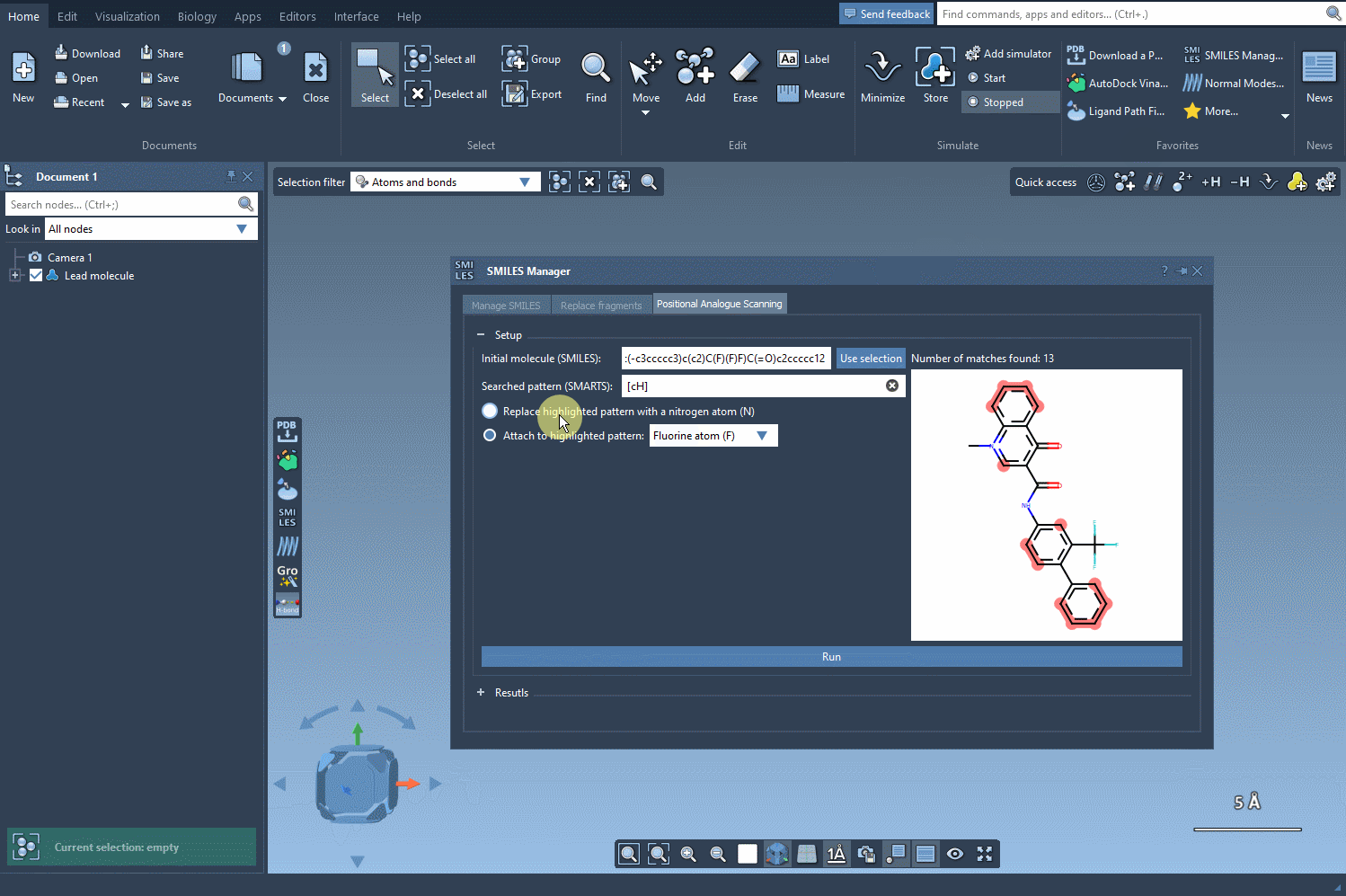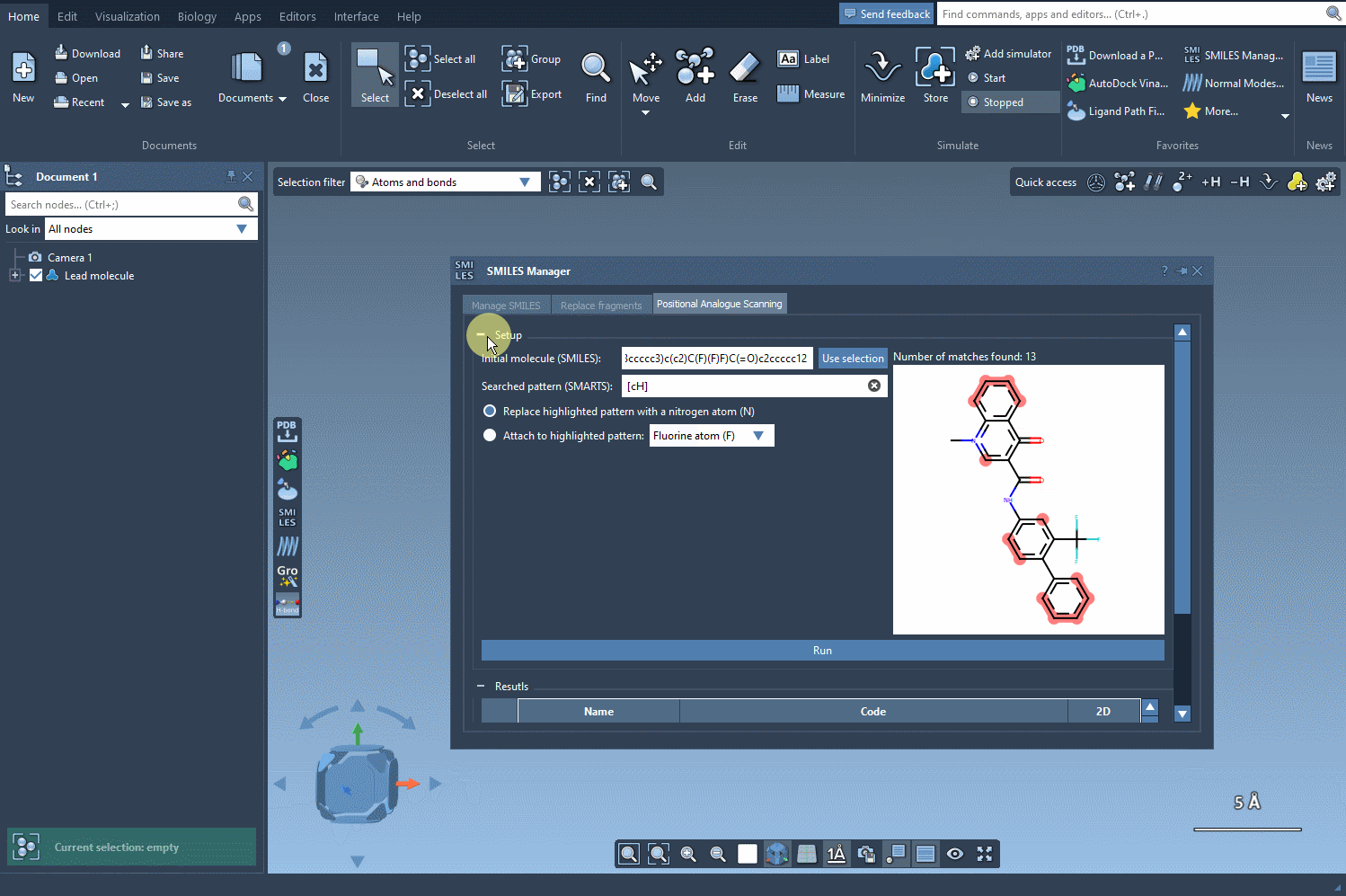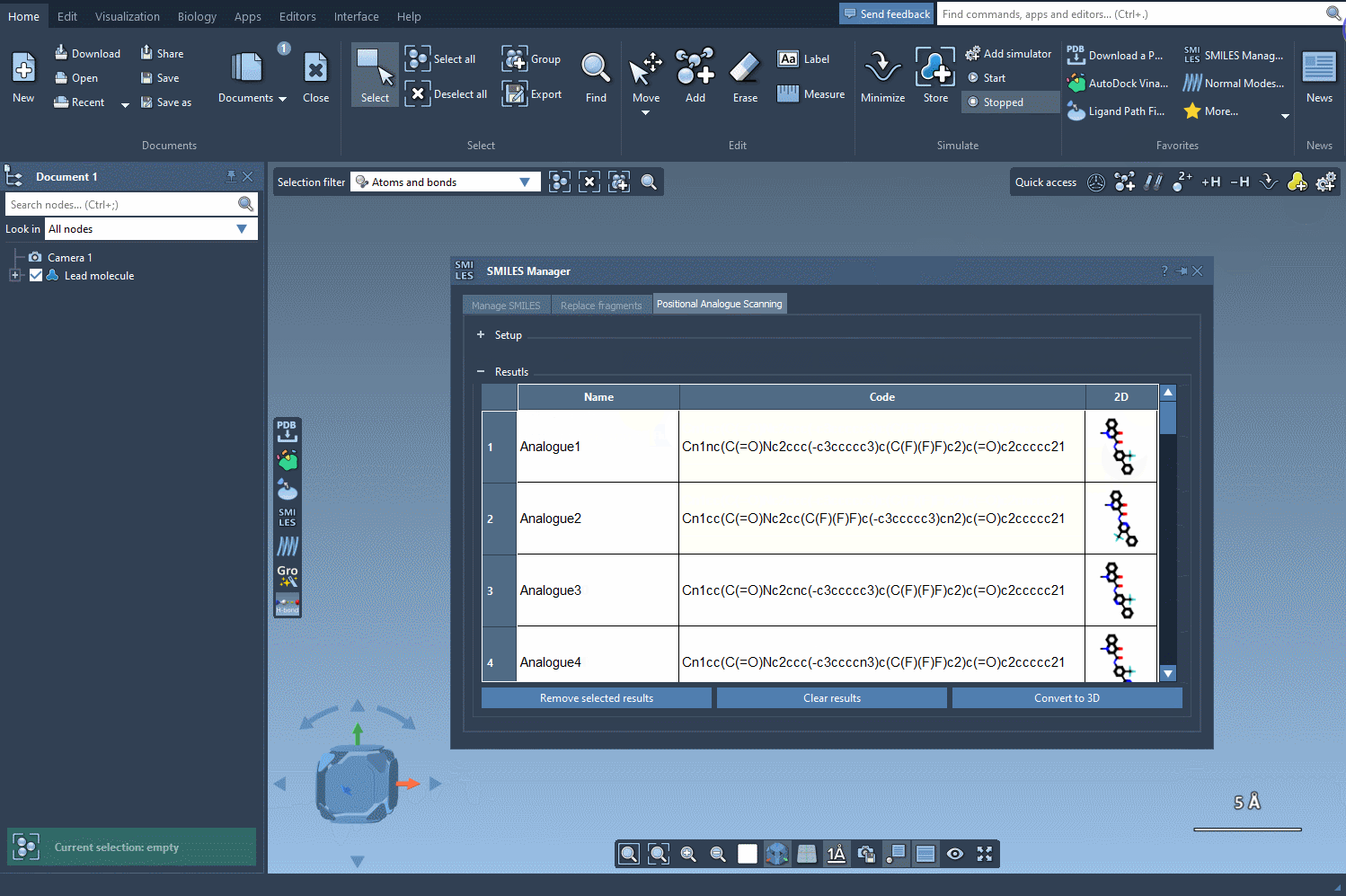
What makes this useful?
This feature is especially helpful for:
- Structure-Activity Relationship (SAR) studies: Systematically vary positions to see what changes improve binding or selectivity.
- Lead optimization: Quickly explore bioisosteres or electron-donating/withdrawing substitutions.
- Teaching and ideation: Visualize molecule changes and educate students or colleagues about chemical properties.
Beyond 2D: Going 3D
Once analogs are generated, you can convert individual or multiple structures to 3D with the Convert to 3D button. This makes them ready for conformational studies, docking simulations, or interaction analysis using other extensions like Autodock Vina Extended.
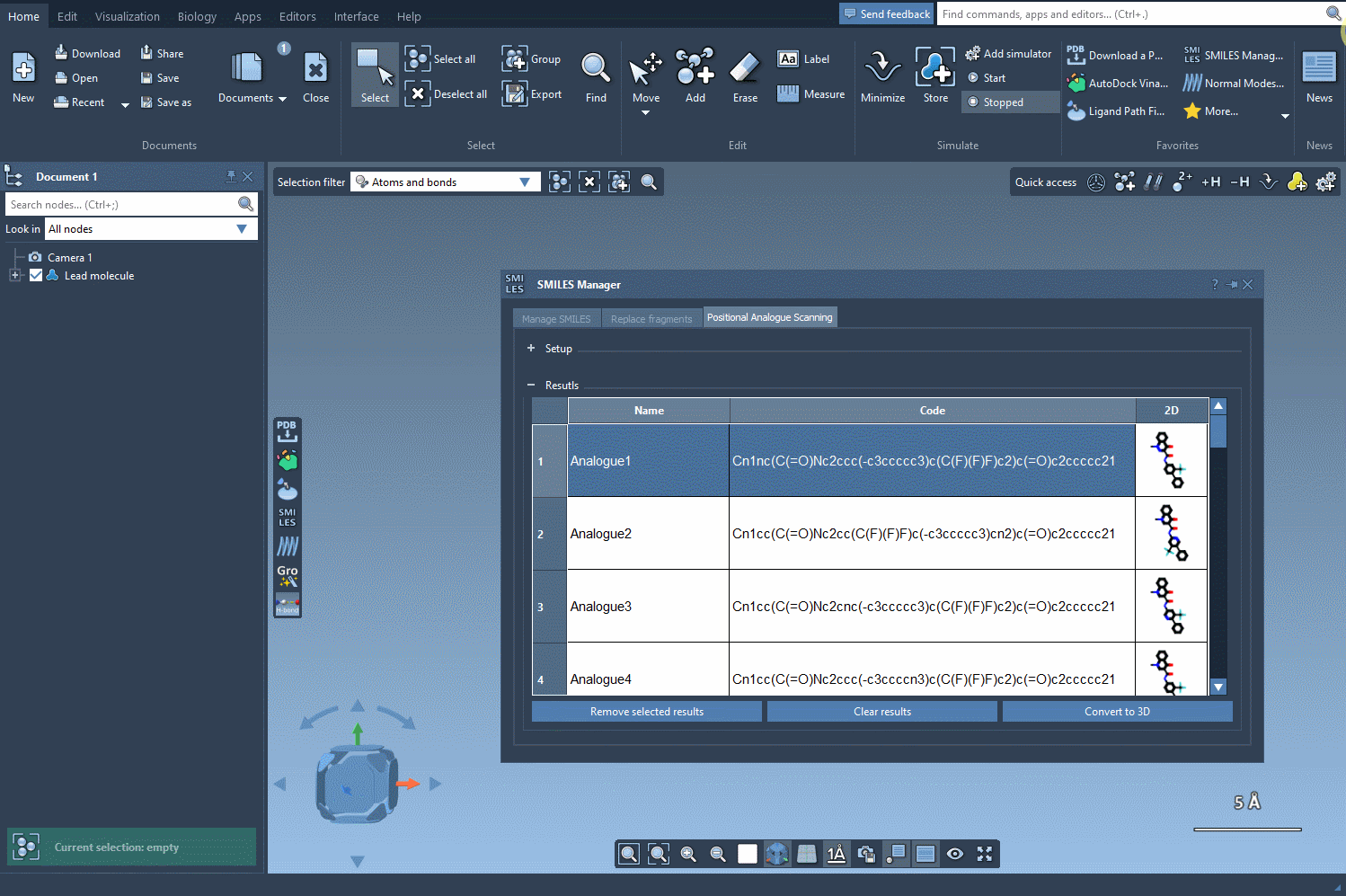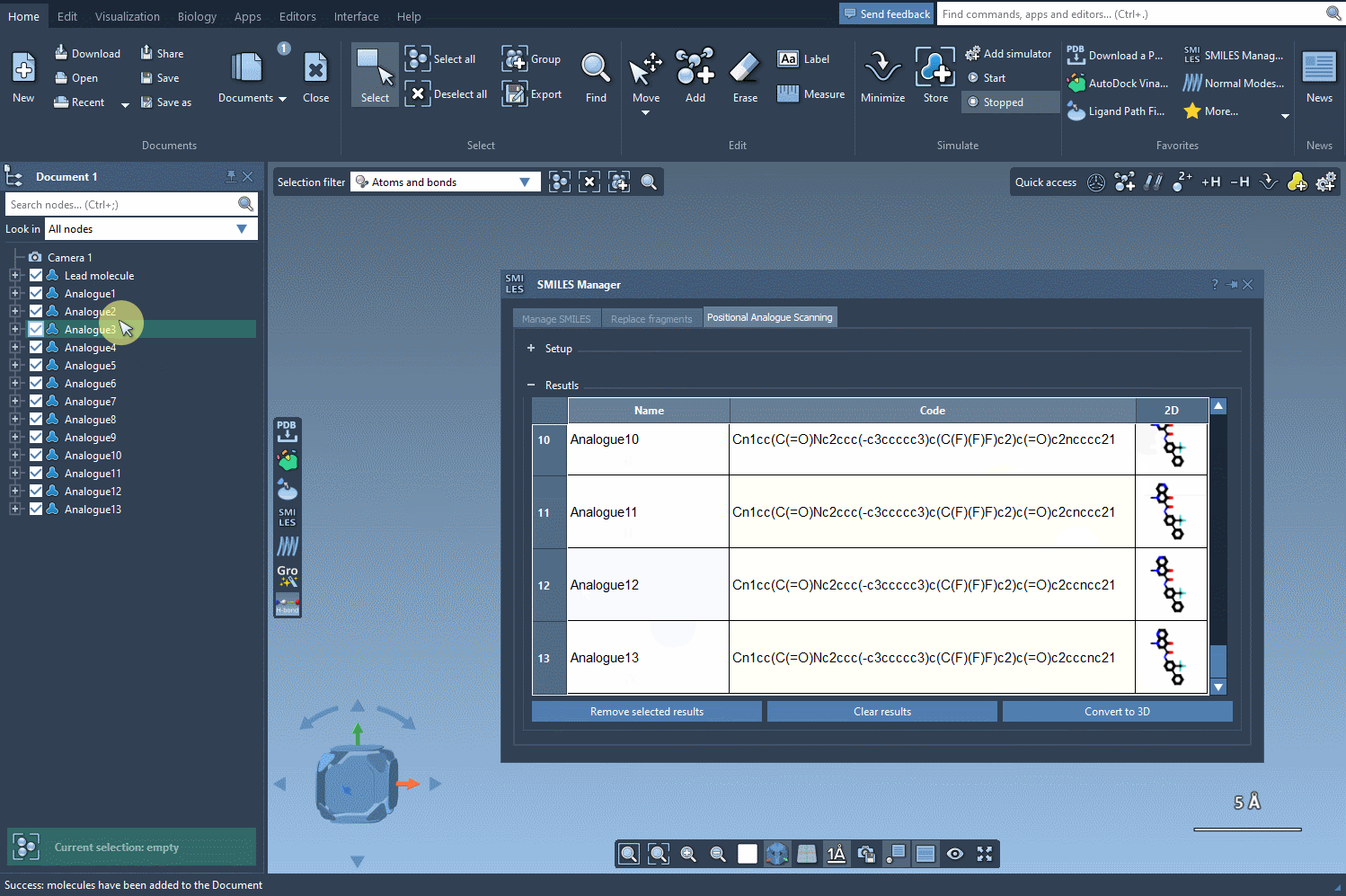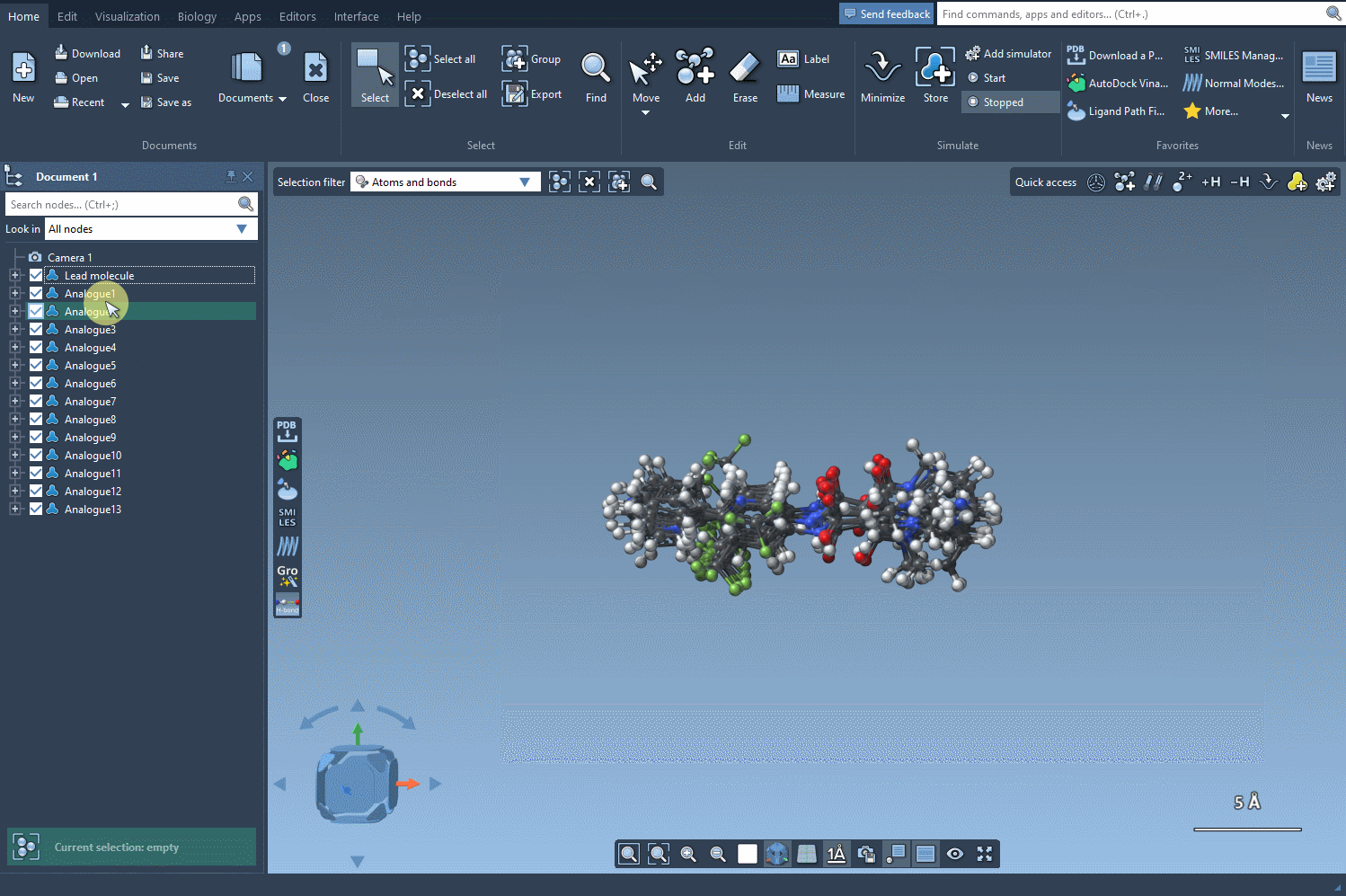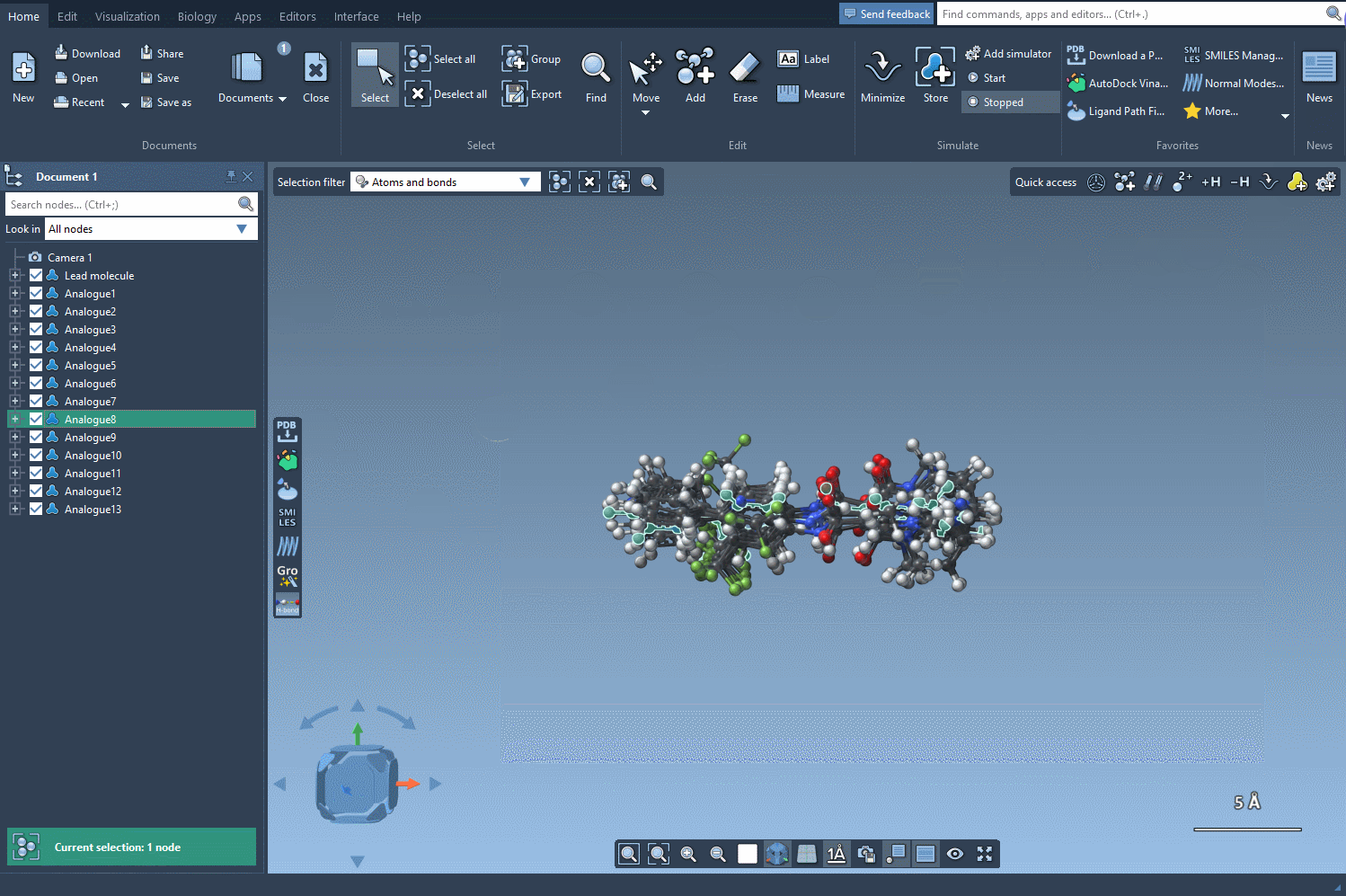
If needed, you can modify names, edit the resulting SMILES, or selectively delete analogs from the result list. The interface supports straightforward interaction, making it easier to filter useful results and avoid clutter.
This ability to move quickly from idea to actual analogs, view structural variations, and prepare them for downstream modeling is a small but powerful part of what SAMSON offers.
Want to learn more about positional analogue scanning and SMILES Manager? Visit the full documentation page here: Positional Analogue Scanning with SMILES Manager.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





