Preparing ligands for docking can sometimes feel like a guessing game: are all hydrogen atoms present? Are rotatable bonds under control? Is the molecule properly minimized? If you’ve found yourself spending more time troubleshooting input data than analyzing results, you’re not alone.
Ligand preparation is one of the most common sources of error in molecular docking. Fortunately, if you’re using SAMSON with the AutoDock Vina Extended extension, there is a clear process to streamline this crucial step. This post breaks down essential tasks and offers tips to avoid common mistakes when getting ligands ready for docking.
📦 Single Ligand or Library?
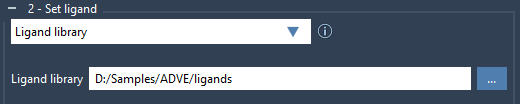
The Vina Extended extension supports docking with either a single compound or an entire directory of ligands. Choose the Single ligand option when dealing with just one molecule. Otherwise, select Ligand library and browse to your folder of ligands in MOL2 or SDF format—even if they are grouped by subfolders.
⚙️ Managing Rotatable Bonds
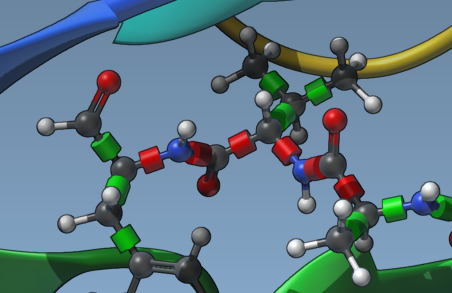
Rotatable bonds play a huge role in determining docking accuracy and performance. When you load a ligand, SAMSON displays green bond controllers in the Viewport to indicate which bonds are rotatable. You can click on these controllers to toggle between rotatable (green) and fixed (red).
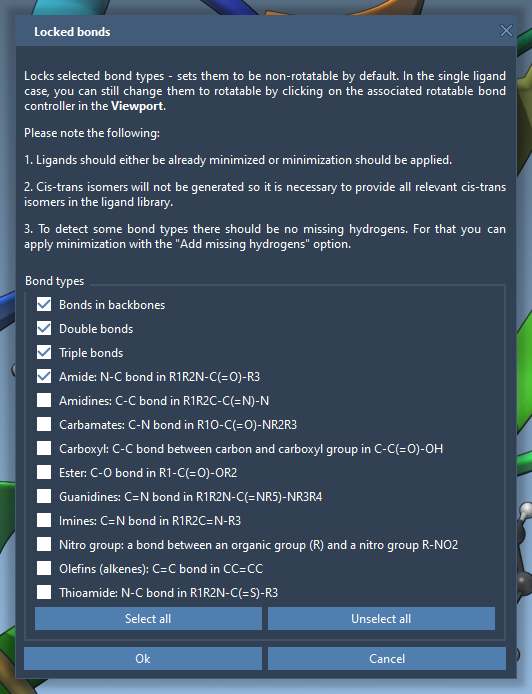
To globally fix certain types of bonds—such as amide or aromatic—you can configure Locked bond settings. This allows for automatic locking of specific bond types during docking, which is particularly useful when processing ligand libraries:
⚛️ Add Hydrogens and Minimize
If your ligands were downloaded from a 2D database or lack hydrogen atoms, check the options Add missing hydrogens and Minimize before docking. Minimization ensures your ligand starts from a physically reasonable conformation, improving consistency across your docking runs.

Choose a minimization preset depending on your needs: more steps yield better-optimized structures but at the cost of speed. Keep in mind that locked bonds won’t restrict minimization—they are only enforced during docking.
🔍 Final Preparation Tip
It’s important to remember that SAMSON does not automatically generate cis-trans isomers. If you need to consider those during docking, you should explicitly include the alternative isomers in your ligand library. This ensures no potential binding modes are missed.
By following these straightforward steps using SAMSON and AutoDock Vina Extended, you’ll save time and avoid many of the most frustrating issues in ligand preparation.
To learn more about ligand setup and related features, including flexible side chains and search spaces, check out the full tutorial on Docking Libraries of Ligands with AutoDock Vina Extended.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON here.





