One of the recurring challenges in molecular modeling is generating plausible intermediate protein conformations between two known states. Whether preparing paths for molecular dynamics simulations or simply aiming to visualize conformational changes, researchers often find themselves lacking tools that strike a balance between speed, realism, and ease-of-use.
If you’re looking for a straightforward way to compute structural transitions in seconds—with control over biological relevance and visualization—SAMSON’s As-Rigid-As-Possible (ARAP) interpolator extension offers a compelling solution.
Why Use ARAP for Protein Transitions?
The ARAP interpolation technique allows users to compute smooth and continuous deformation paths between two protein structures. This can serve multiple purposes:
- Generating input trajectories for umbrella sampling, steered MD, or P-NEB techniques.
- Exploring structural hypotheses during conformational analysis.
- Improving presentations and communication using visual interpolations.
Use Case: Transition from 1DDT to 1MDT (Diphtheria Toxin)
Let’s walk through a common use case: generating a path from chain A of 1DDT to chain A of 1MDT, two conformations of the Diphtheria Toxin.
Step 1: Load and Clean the Structures
- In SAMSON, go to Home > Fetch and input
1DDT 1MDT. - Delete unwanted chains (like chain B in 1MDT).
- Run Home > Prepare to remove all waters, ligands, and alternate positions.

Step 2: Create Conformations
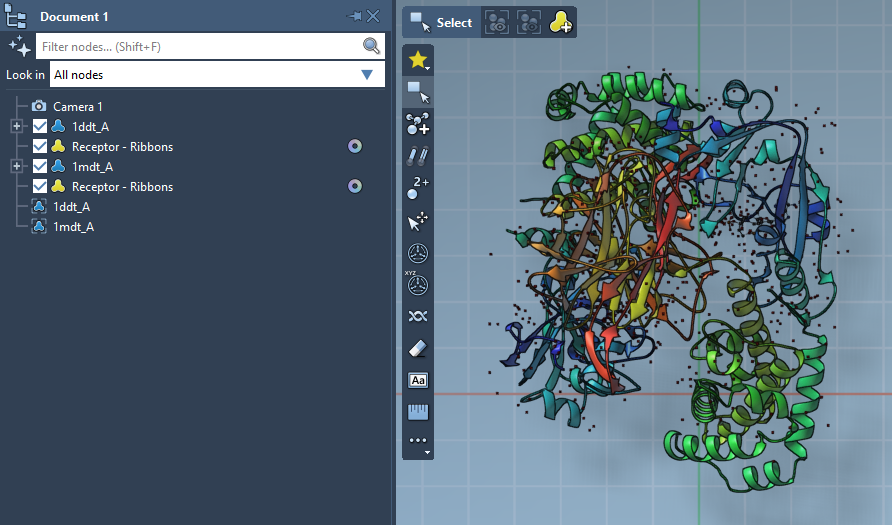
- Select each cleaned structure in the Document View.
- Go to Edit > Conformation and name the conformations, e.g.,
1DDT Aand1MDT A.
These conformers define the start and goal states of your transition path.
Step 3: Configure and Run the ARAP Interpolator
- Access the interpolator app via Home > Apps > Biology > ARAP Path Interpolation.
- Use Get conformations from active document and select start =
1DDT A, goal =1MDT A. - For atom matching, choose All except hydrogens.
- Construct edges from bonds and optionally connect α-carbons across missing residue segments.
- Enable Perform alignment and set number of path conformations to
20.
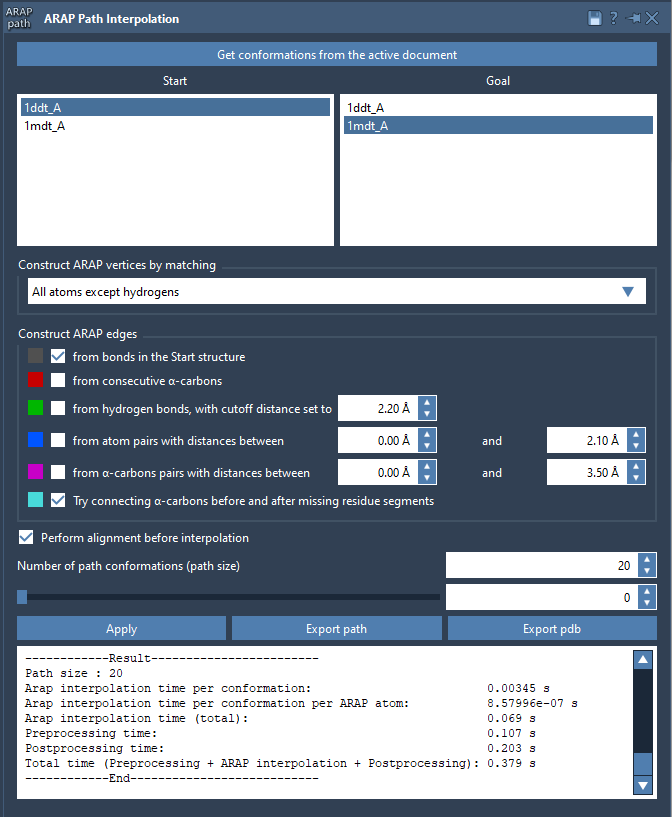
Click Run and within seconds you’ll get the complete set of interpolated conformations.
Visualize and Export Your Results
Use the slider to scrub through the transition and observe intermediate structures in real-time:
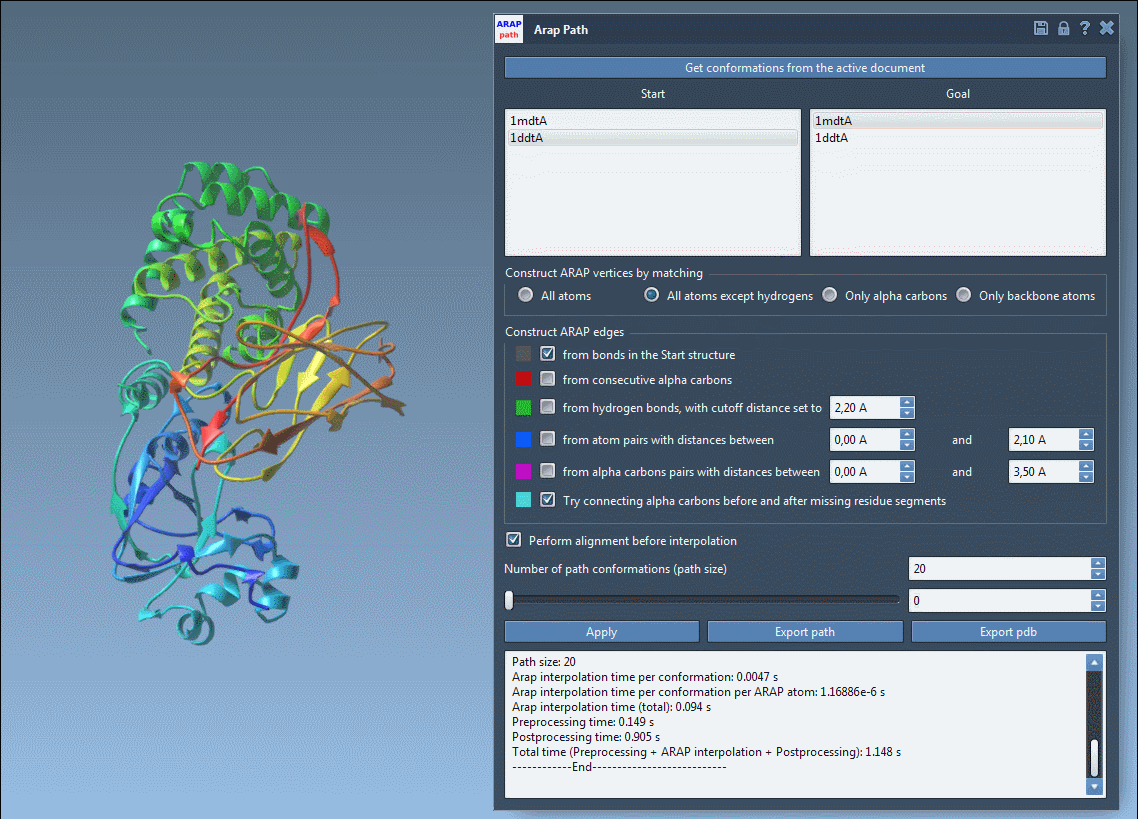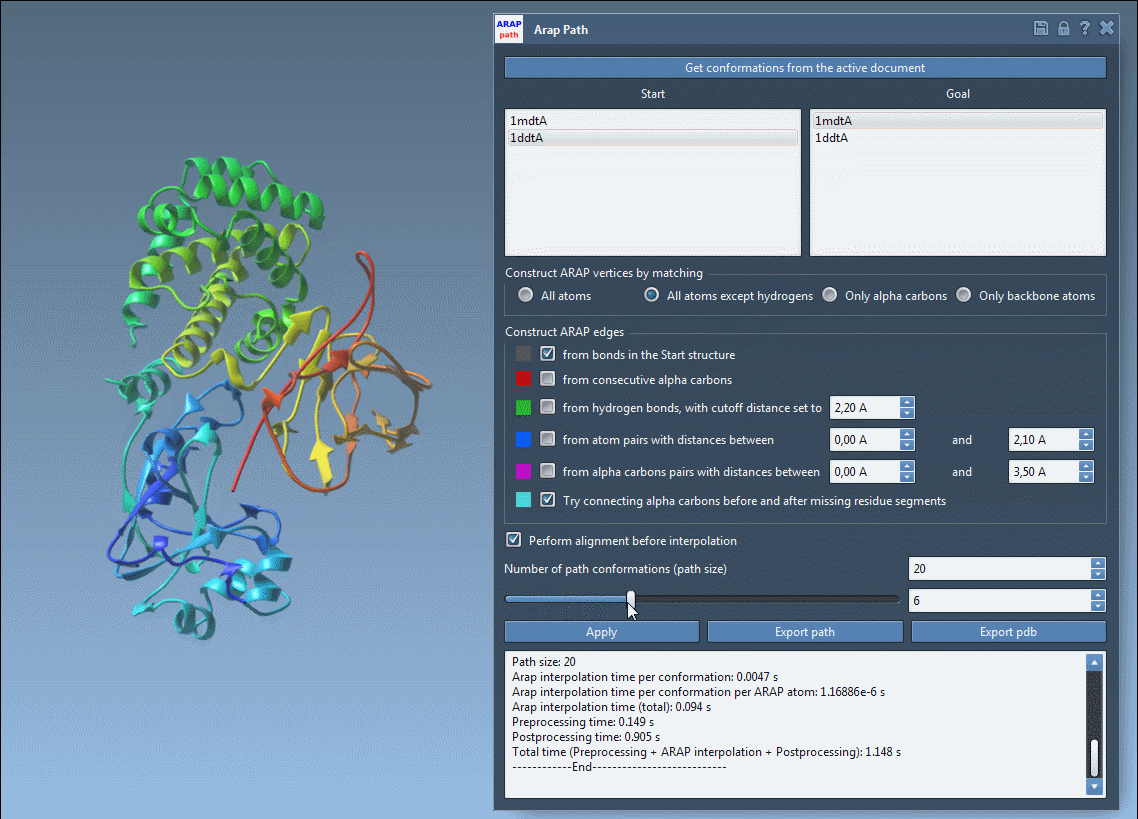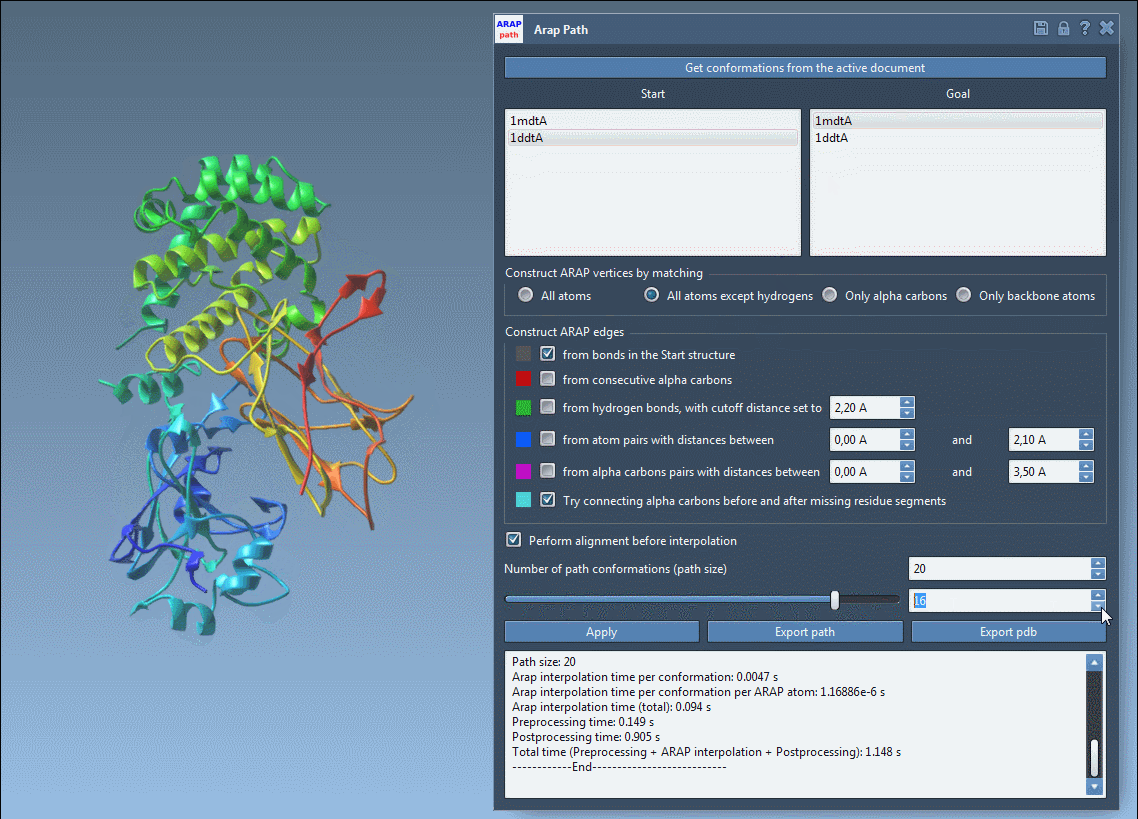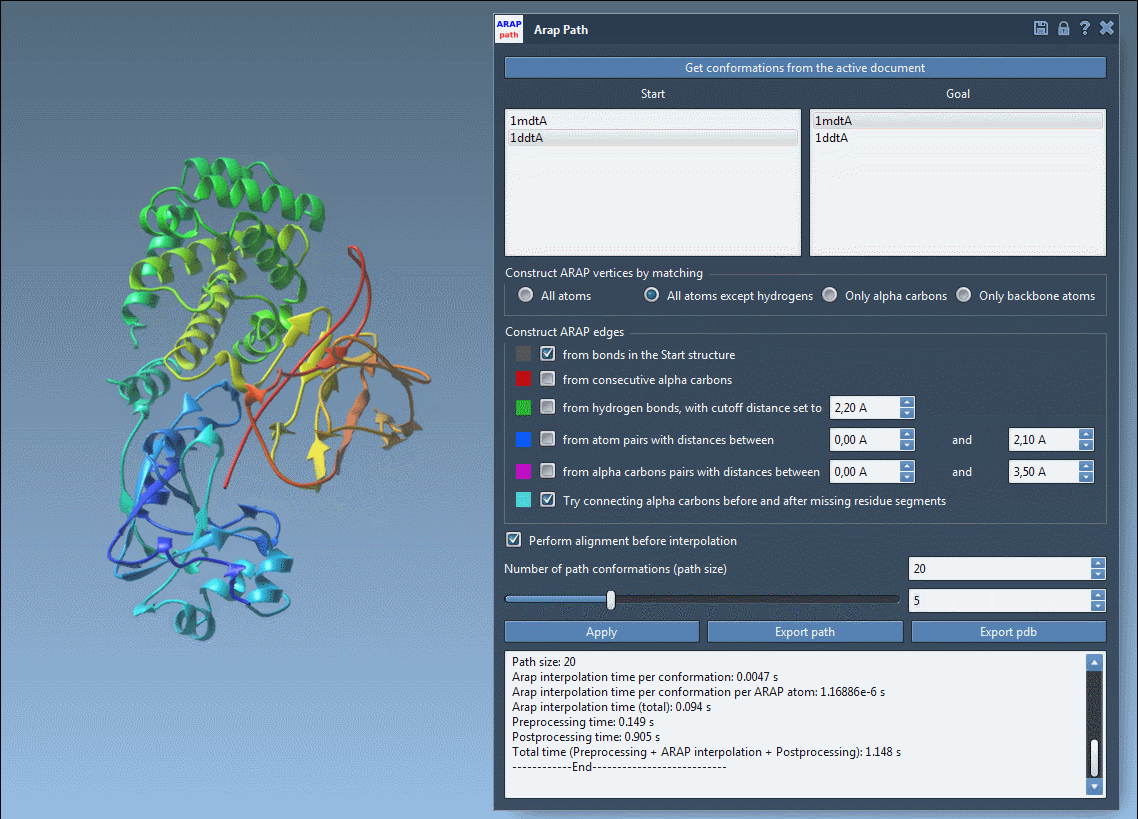
- Visualize ARAP edges to understand structure connectivity.
- Export as trajectory or PDB series for downstream use in simulations.
Tip: Avoid Common Errors
If you encounter the error: “structure does not make one connected component”, revisit the Prepare step to ensure you’ve removed water, ions, and ligands.
Where to Go from Here?
The resulting ARAP path can be paired with simulation tools like GROMACS Wizard for umbrella sampling or refined using P-NEB. It also serves well for dimensionality reduction workflows or to produce clearer structure animations in presentations or manuscripts.
To learn more about configuring ARAP interpolation and other use cases, visit the full documentation: ARAP Interpolation for Protein Structures.
SAMSON and all SAMSON Extensions are free for non-commercial use. Download SAMSON at www.samson-connect.net.





