When designing new molecules, one recurring challenge is to understand how small chemical changes affect biological activity or interactions. Whether you’re optimizing binding affinity, reducing toxicity, or simply exploring chemical space, one question often arises: what happens if I slightly change this part of the molecule?
A straightforward approach to this problem is positional analogue scanning: systematically replacing or modifying individual atoms or groups in a molecule to analyze how each change impacts the molecule’s behavior. This can lead to new insights—and potentially better drug candidates.
Luckily, using the SMILES Manager extension in SAMSON, this process becomes visual, interactive, and fast.
Scanning a molecule step by step
Let’s take a closer look at how you can start generating your own molecular analogues in just a few steps using positional analogue scanning inside SAMSON.
Step 1: Define your molecule
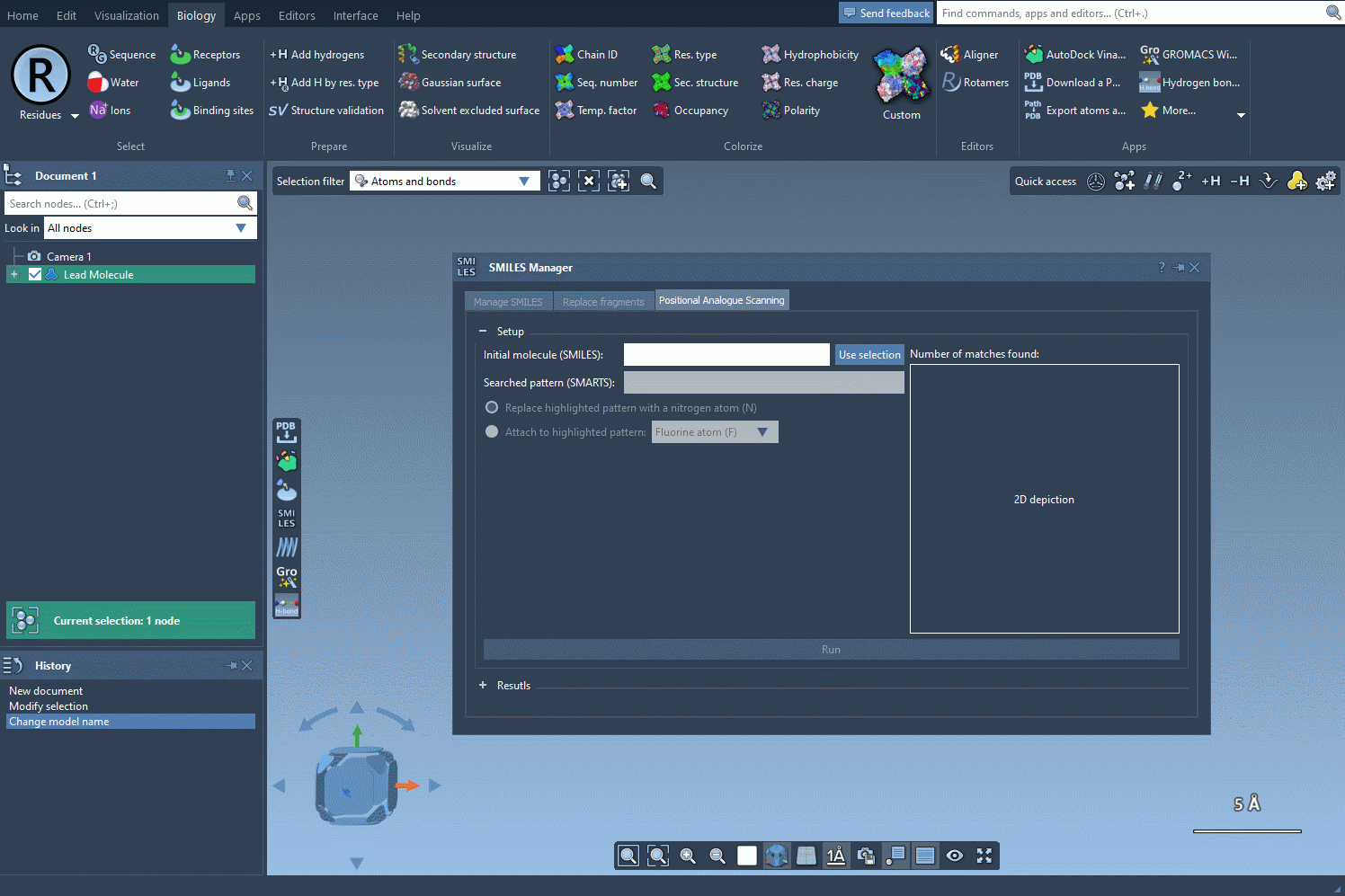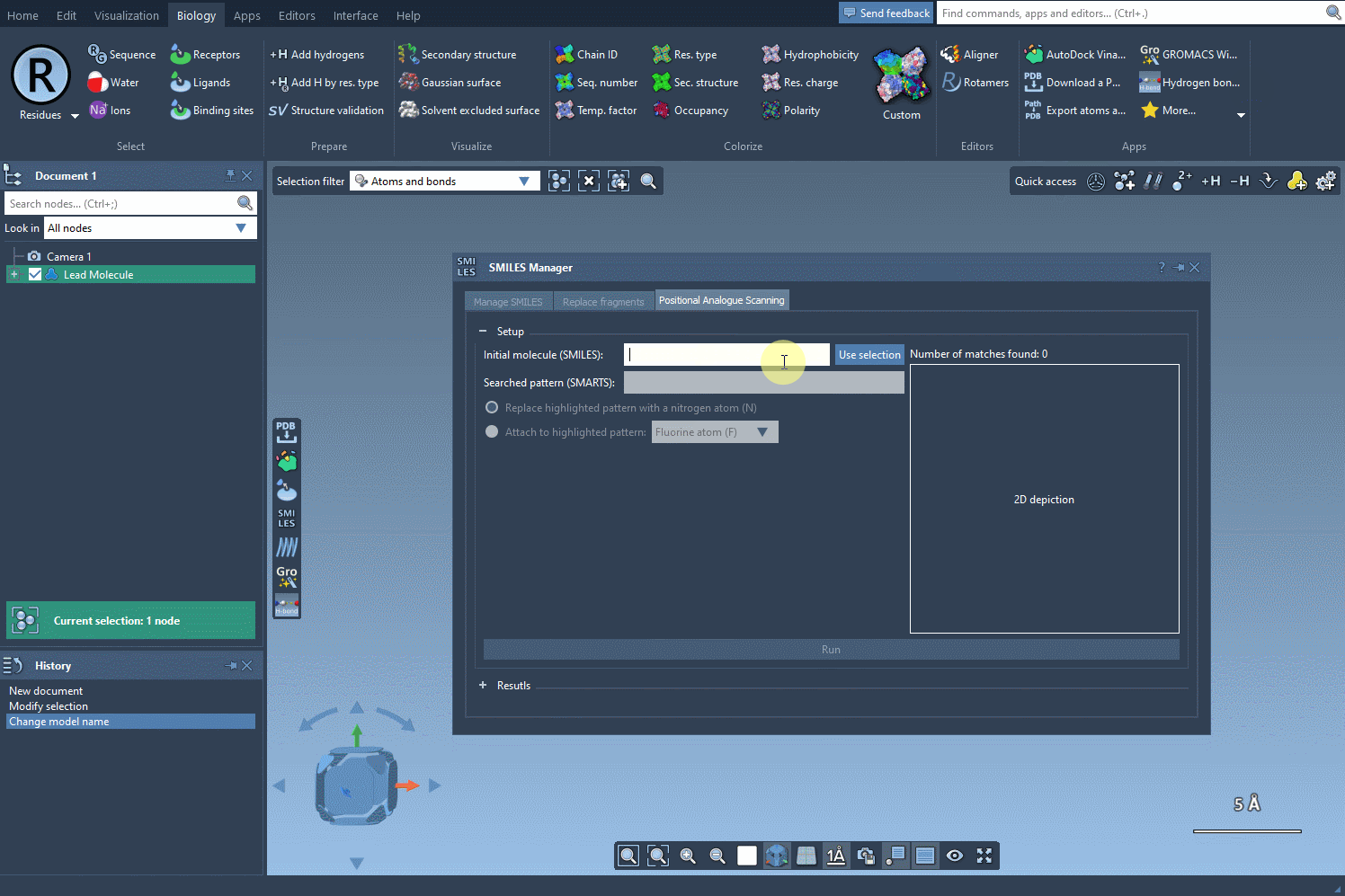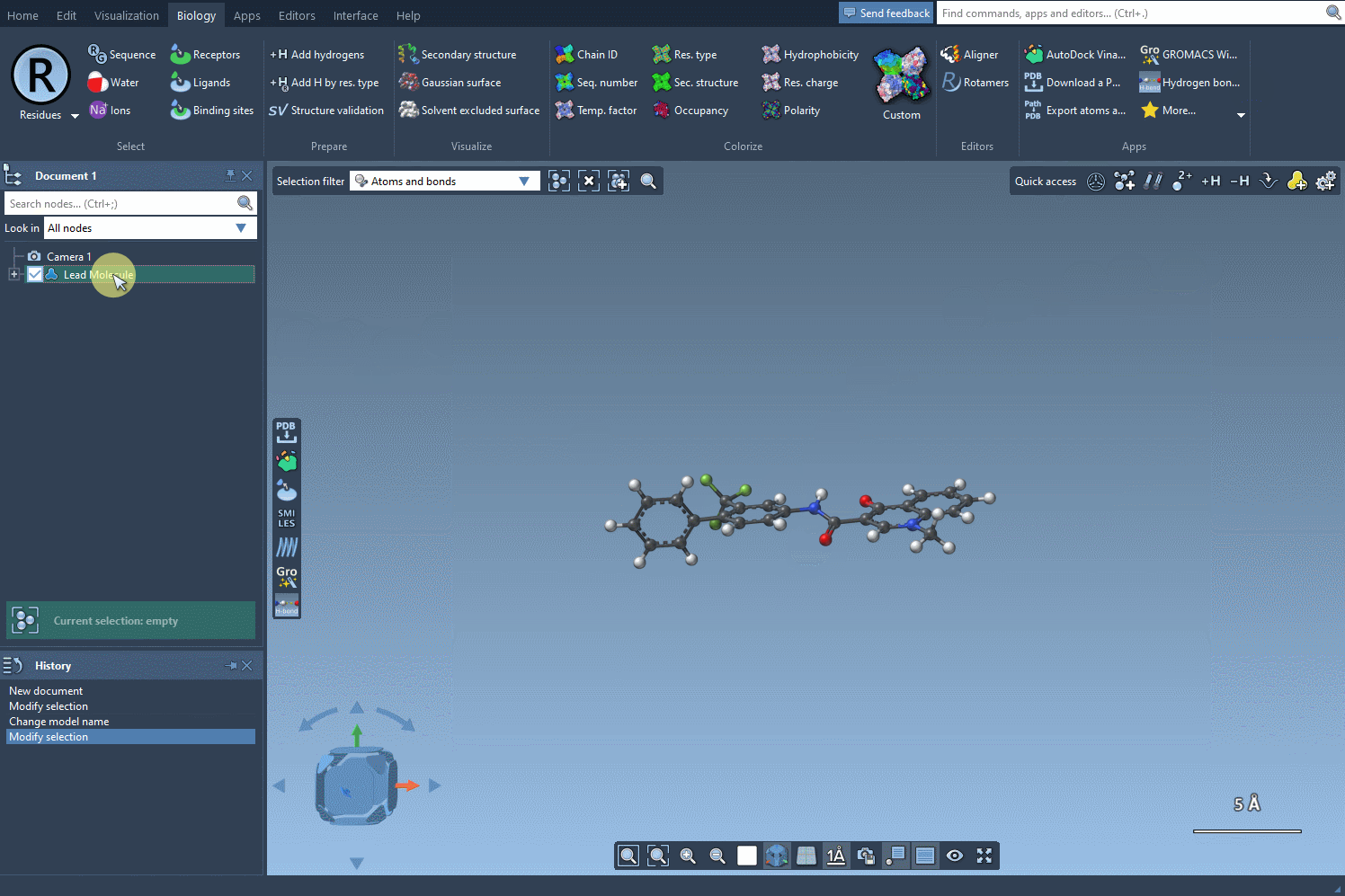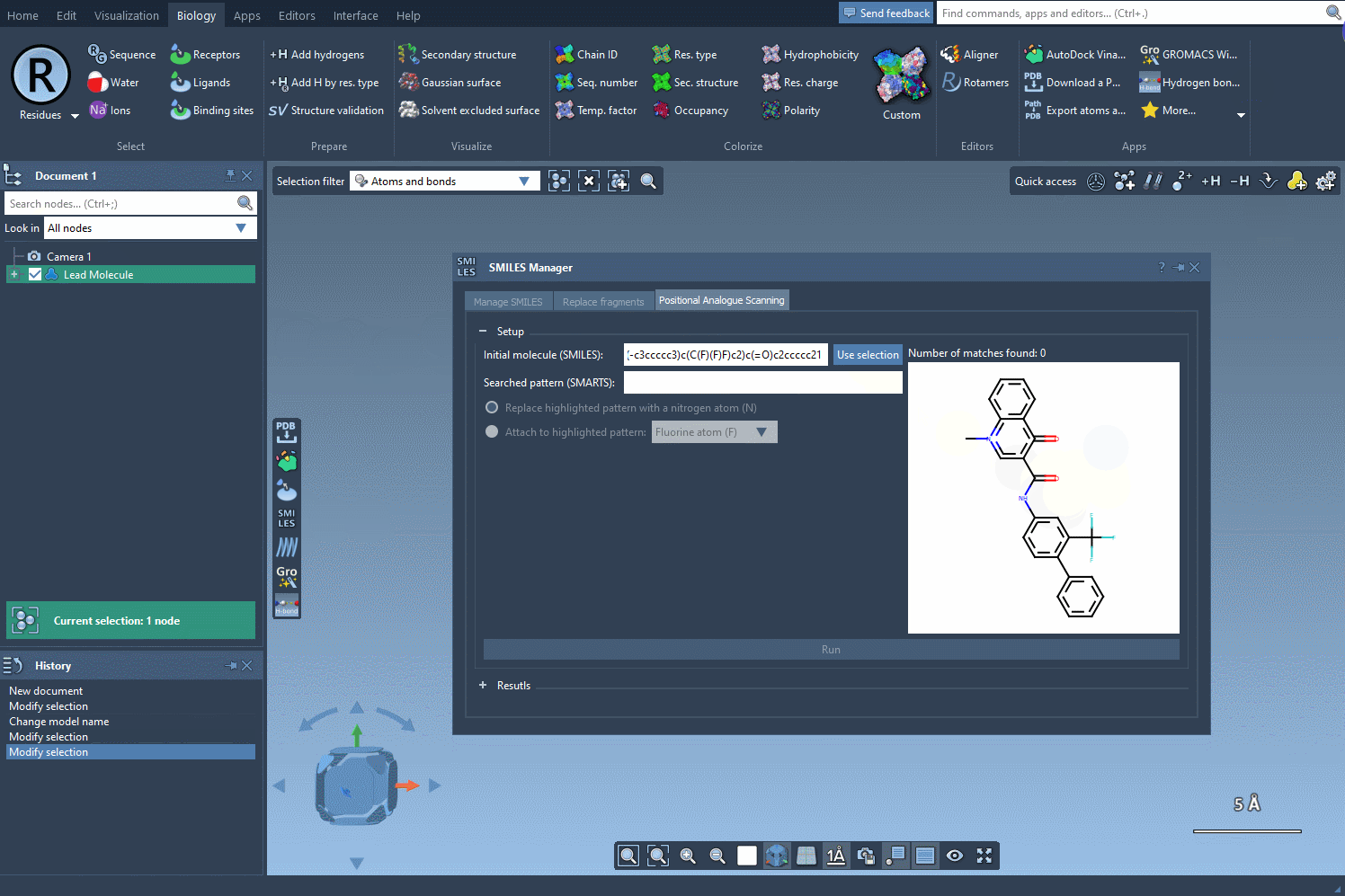
Everything begins with a molecule. You can either paste a SMILES string or select an existing molecule in your SAMSON document. A button labeled Use selection makes the process seamless and intuitive—for example, using a reference molecule from a published study.
Step 2: Highlight a pattern to modify
What do you want to change in the molecule? An atom? A group? Using SMARTS patterns, you can specify which substructures to target. For instance, entering the pattern [cH] will search for aromatic carbon atoms with a single hydrogen. Once entered, the matching regions are automatically highlighted in the molecular structure.

Step 3: Swap or expand
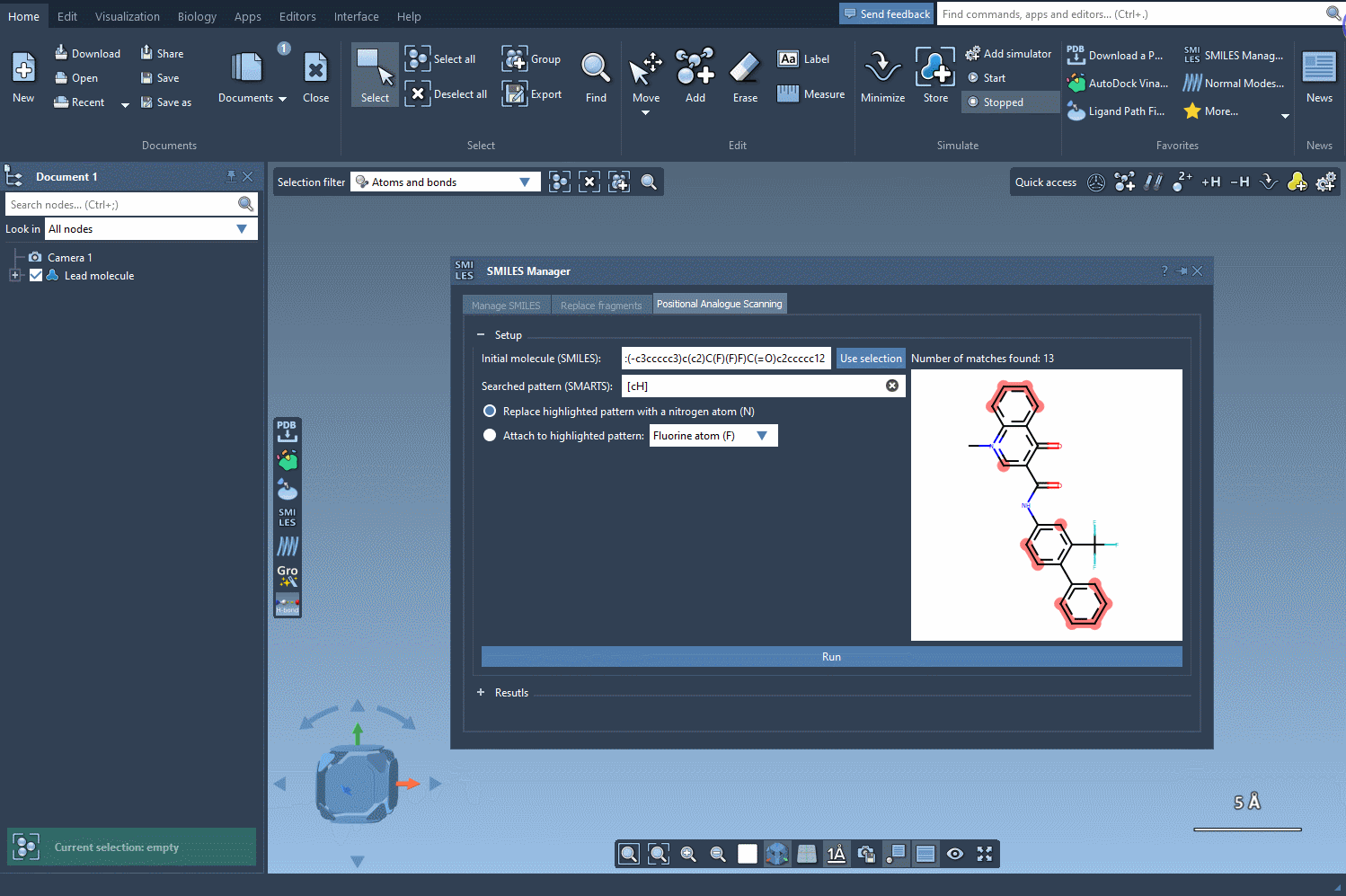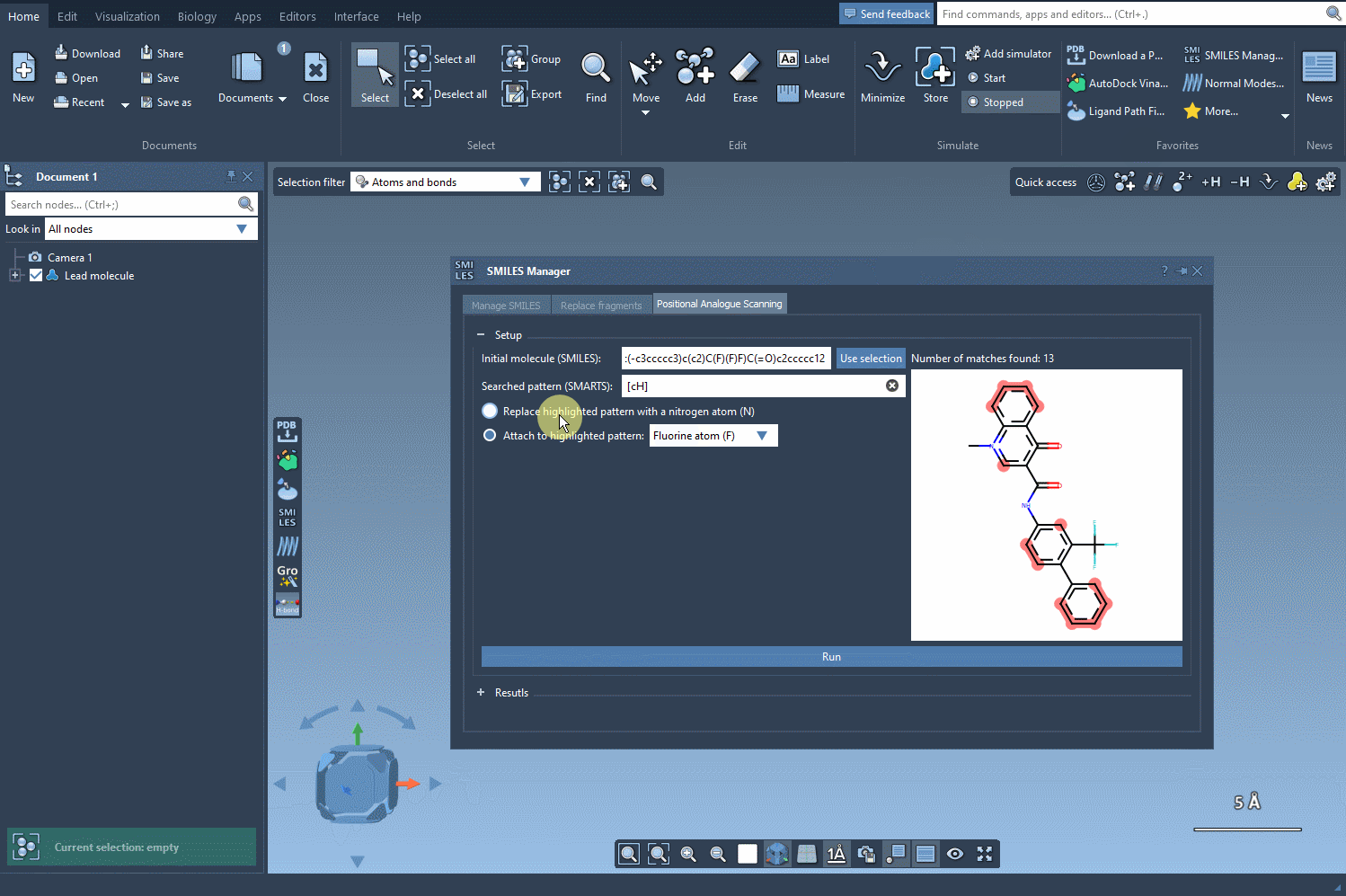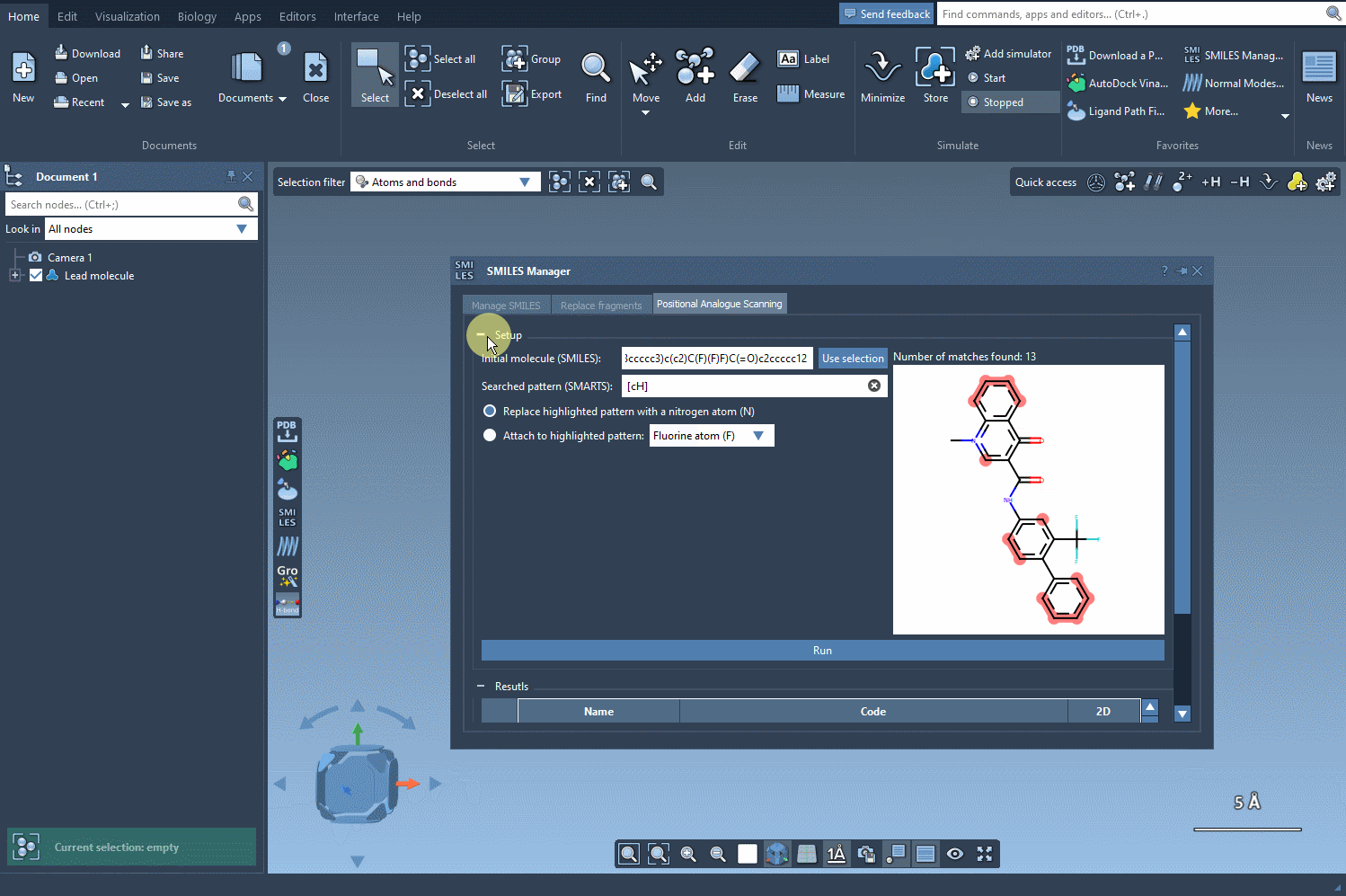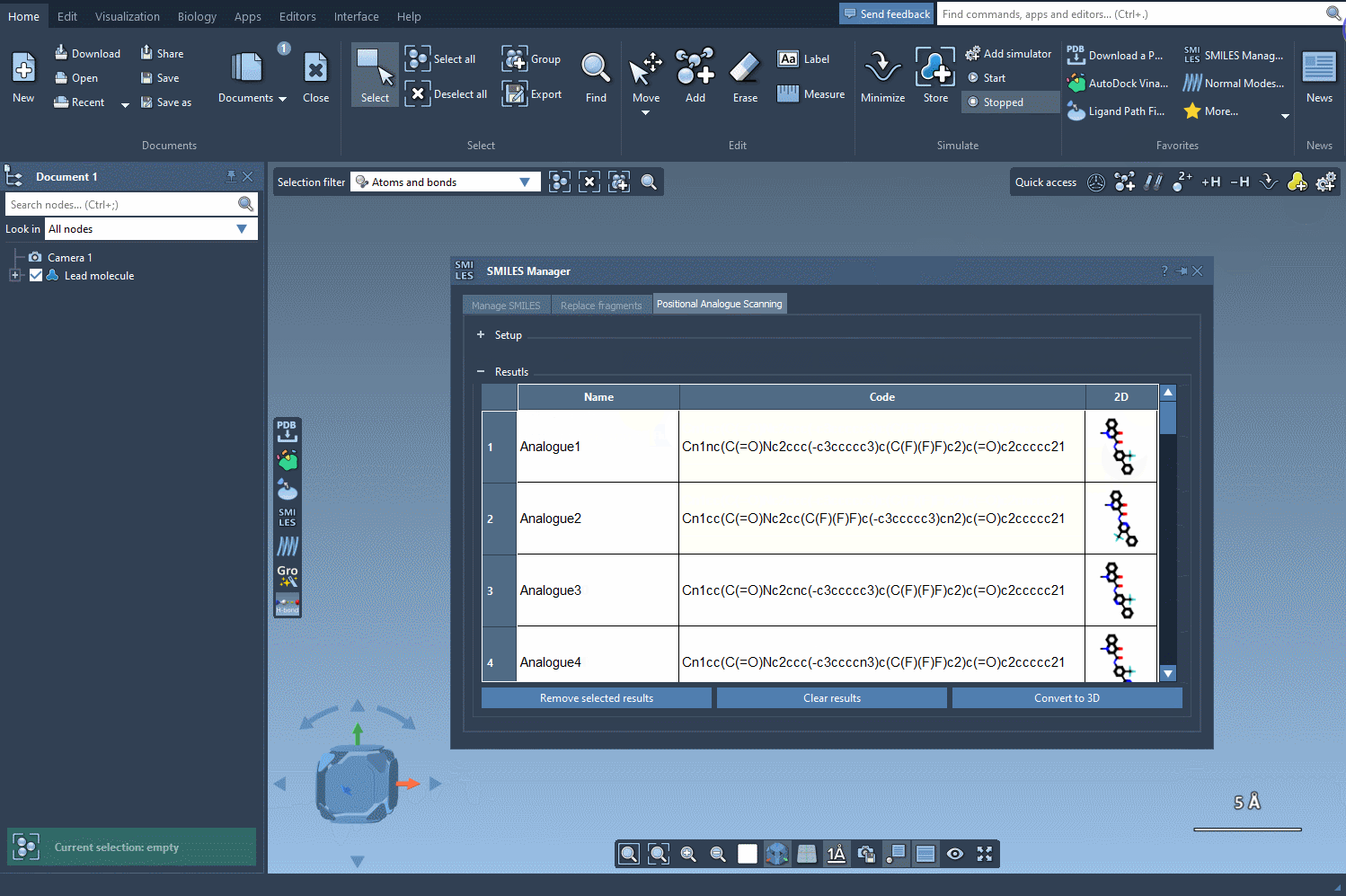
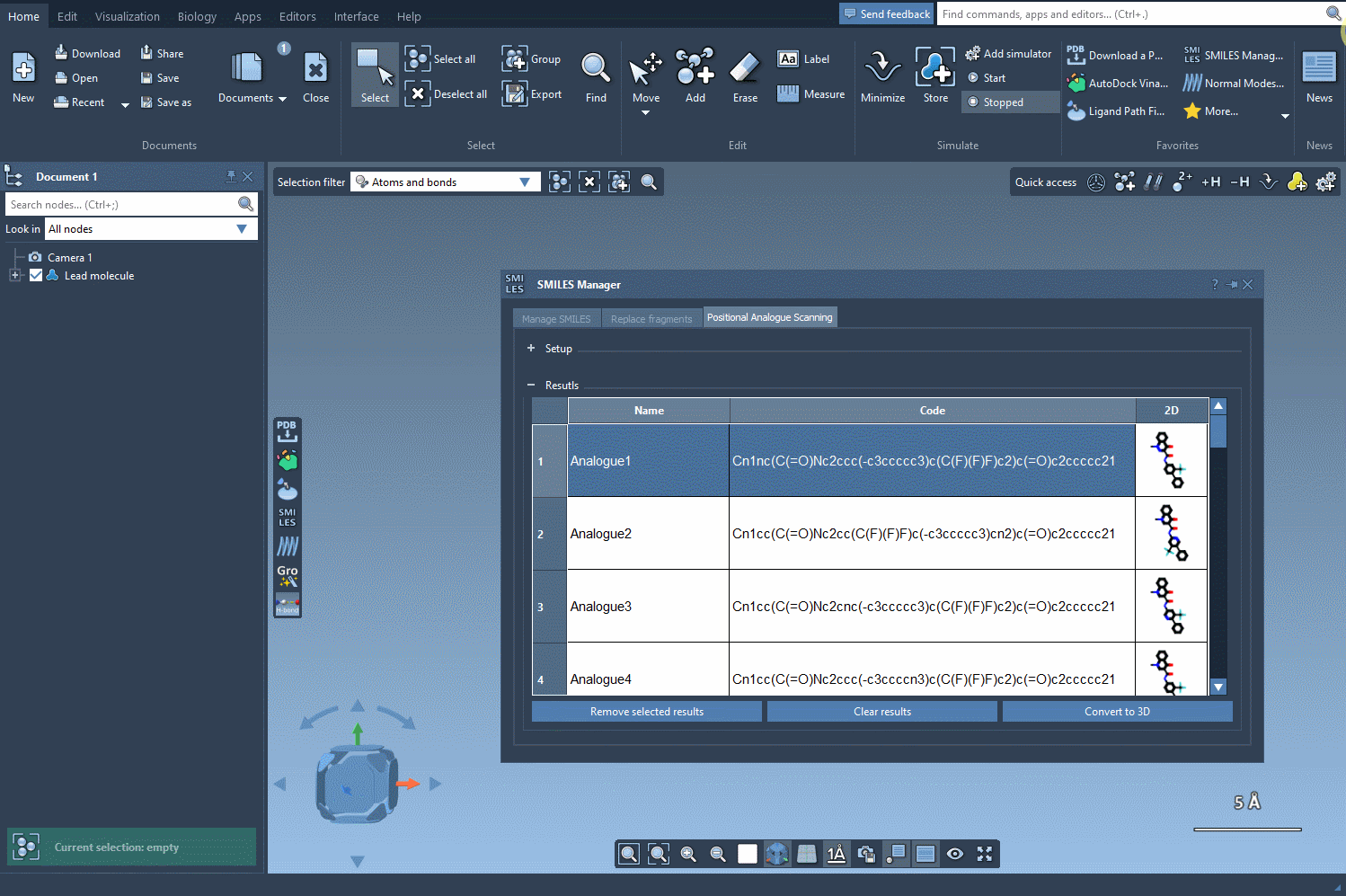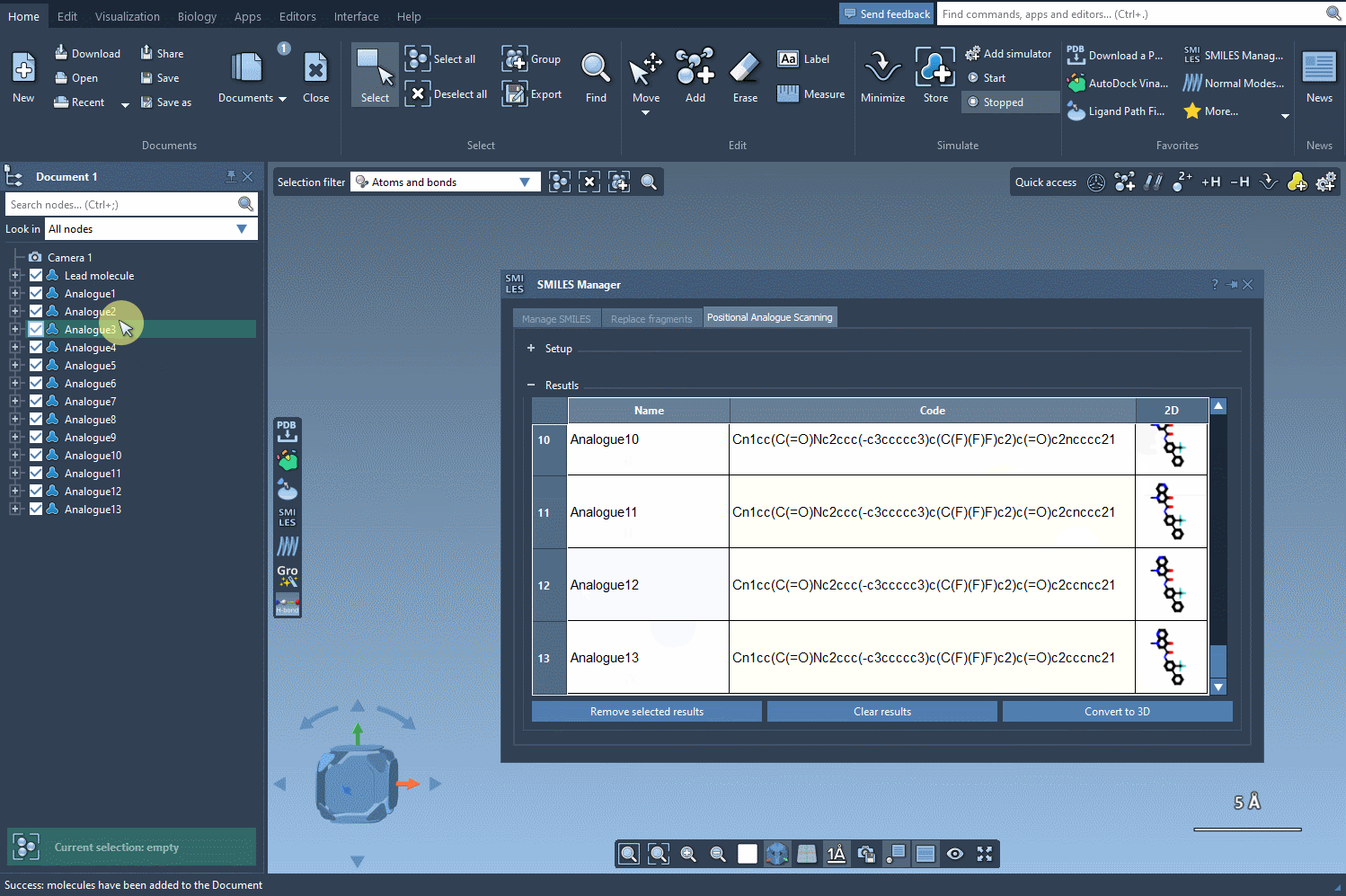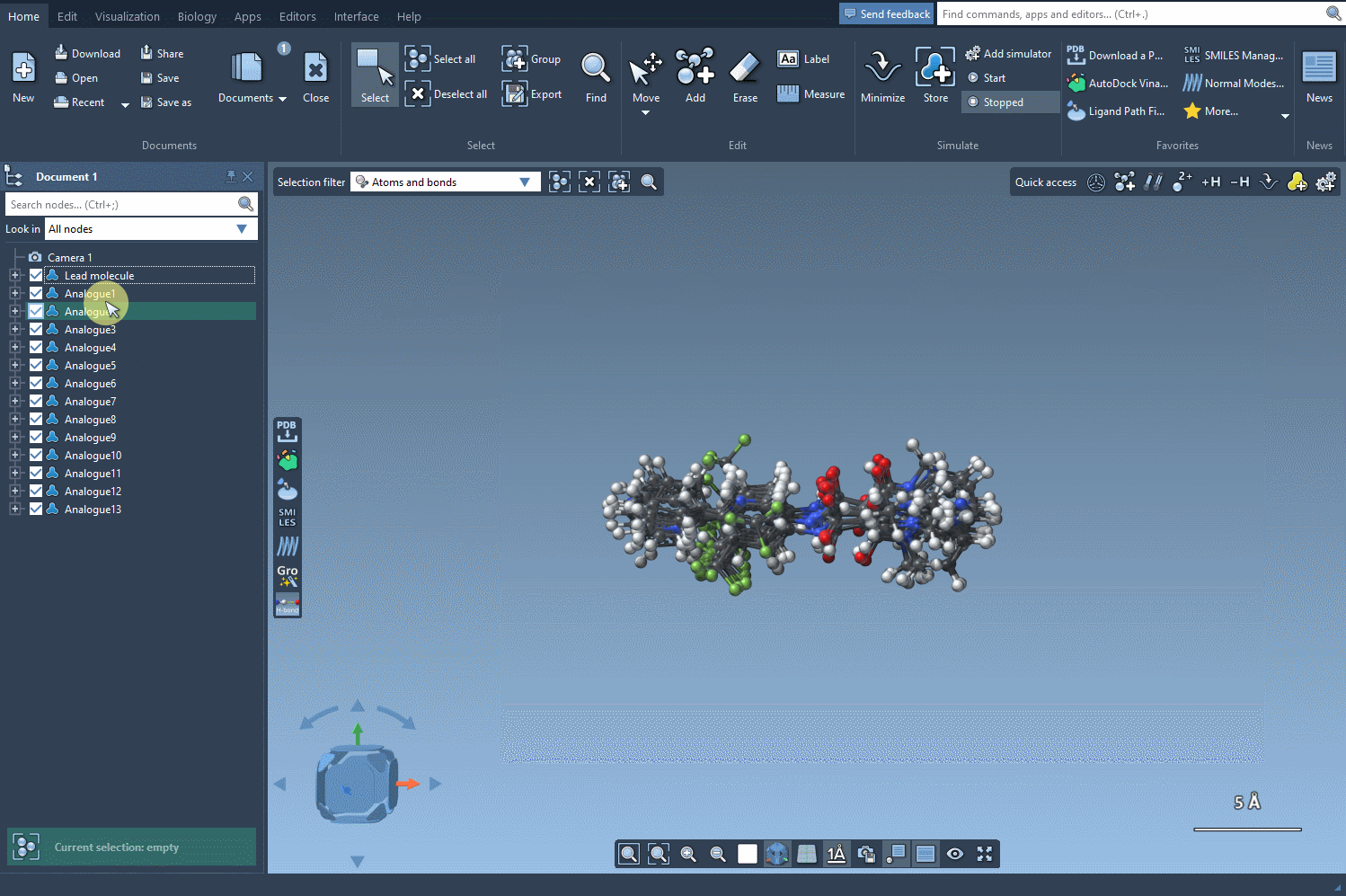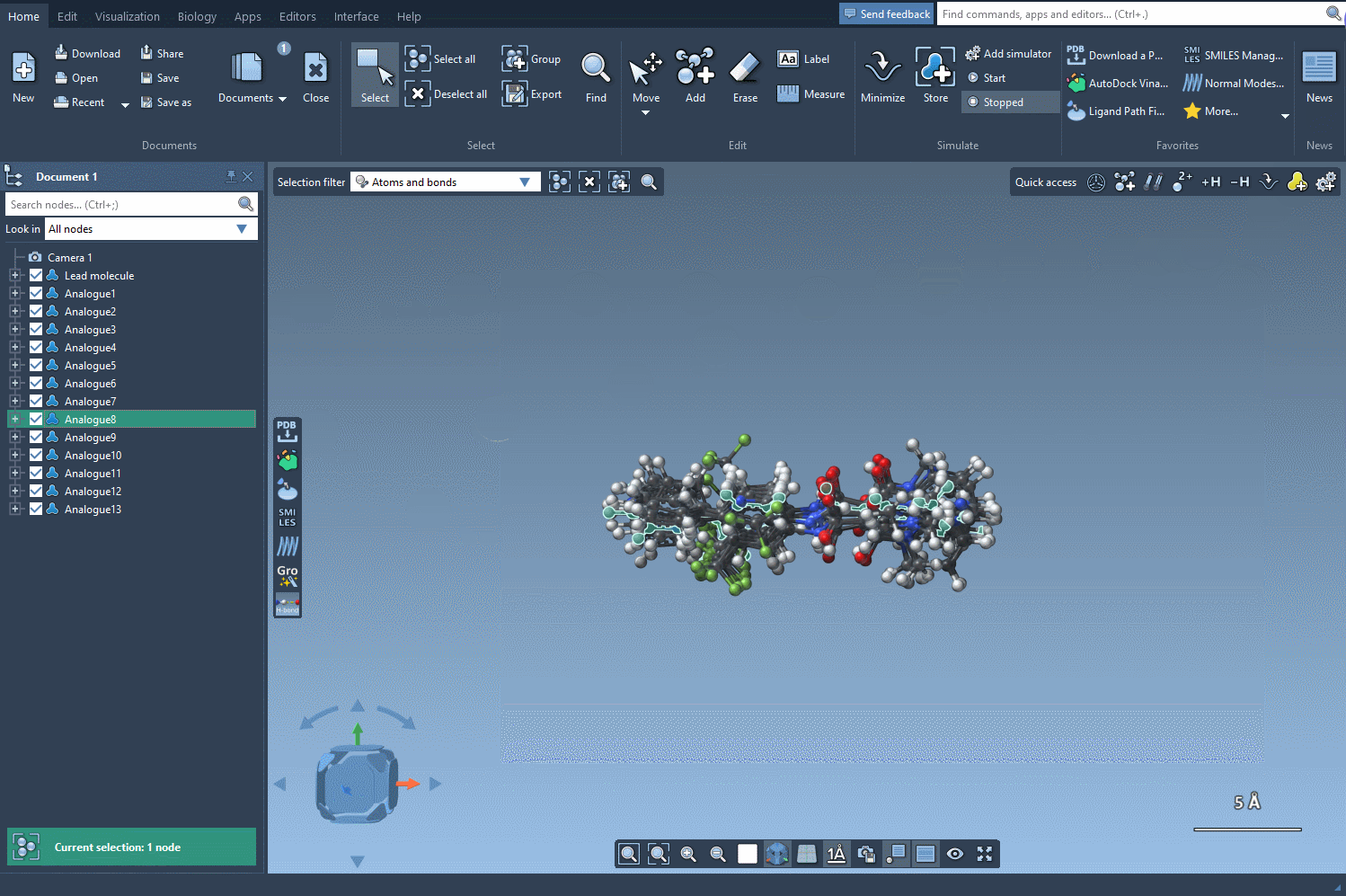
Now comes the transformation. You can choose to replace the selected pattern (e.g. swap a carbon for nitrogen) or attach new functional groups like fluorine or methyl. Click Run and the SMILES Manager quickly generates a list of analogues that reflect your specified changes. Each result includes the SMILES string and a 2D image for easy navigation.
Step 4: Modify, inspect, iterate
You’re not stuck with the default results. Modify individual analogue names or their SMILES codes directly in the results table. Need more space? Double-click the 2D image to expand it. Want to remove just a few results? You can do that too. The tool is flexible and designed to let you explore as you go.
Step 5: Build the 3D version
Once satisfied with your 2D analogues, convert them to 3D using the Convert to 3D button. These 3D models can now be used for docking studies or visual inspection inside SAMSON to evaluate interactions with target proteins.
Why this helps
Positional analogue scanning is not just about chemistry—it’s about insight. Understanding which atomic changes impact binding or stability helps guide experimental decisions. Instead of guessing what might work, you can prioritize changes based on clear trends observed in silico. The visual and interactive nature of SAMSON’s SMILES Manager makes this process more accessible and efficient for molecular modelers at all levels.
Want to try it yourself? Explore the full tutorial here.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





