When studying molecular interactions, understanding how specific atoms move along a reaction or unbinding pathway can reveal critical insights. Whether you’re preparing reaction coordinate input files for enhanced sampling, or trying to isolate ligand movement for free energy calculations, exporting only the relevant atoms from an MD or path-based simulation is a common roadblock.
Luckily, SAMSON’s Export Along Paths Extension provides an efficient solution to track and export the trajectory of a subset of atoms—like a ligand—along predefined paths. Here’s how you can use it to your advantage.
Select Only What Matters
Instead of writing out full trajectory files for entire molecular systems, this method allows you to target exactly what you need—ideal for reducing file size, decluttering data, and honing in on binding/unbinding dynamics.
Step-by-Step: Exporting a Subset of Atoms (e.g., a Ligand)
- First, open the Export Along Paths app in SAMSON via
Home > Apps > All > Export Along Paths. - In the Document view, select the subset of atoms you want to track. For this tutorial, let’s use the ligand
TDGas an example.

- In the app’s interface, expand the Advanced panel, and click Add to define the selected atoms as a new model for export.
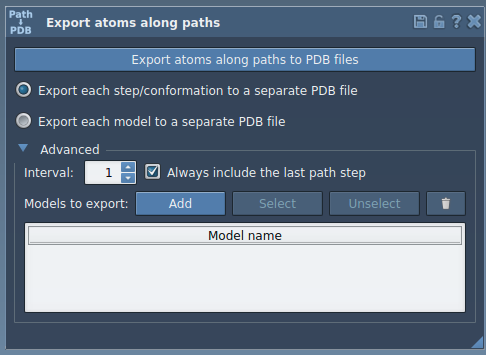
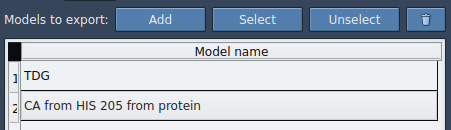
This will add the selected group to an export table. You can rename the group, select/unselect atoms for visual confirmation, and manage multiple atom groups for simultaneous export.

Multimodel Support
You can define several models (e.g., ligand + catalytic site), export them simultaneously, and organize your trajectory exports accordingly.
Final Export
Once you’ve defined your atom groups:
- Select the export mode (single or multiple PDB files).
- Choose the relevant path(s) in the Document view.
- Click Export atoms along paths to PDB files.
The tool prompts you to choose the export destination and file name prefix.
Why This Matters
Whether you’re assembling files for umbrella sampling, visualizing intermediate states, or preparing input for free energy calculations, being able to extract just the ligand or critical regions makes your workflow faster and cleaner.
This selective export reduces post-processing time and is particularly useful when working with large biomolecular complexes where only a small region is under study.
To learn more about exporting atom trajectories along paths in SAMSON, visit the full documentation here:
https://documentation.samson-connect.net/tutorials/export-along-path/export-atoms-trajectories-along-paths/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





