Creating realistic membrane-protein systems is a recurring challenge in molecular modeling. Membrane proteins play critical biological roles, but setting up simulations that combine them with appropriate lipid environments often involves multiple tools, file format conversions, and a fair amount of scripting.
The Molecular Box Builder extension in SAMSON offers a more streamlined approach. It allows you to rapidly generate lipid layers or bilayers around proteins directly in your molecular design environment. This post walks you through the exact steps to go from a bare protein to a lipid bilayer system, ready for further refinement or simulation — no external tools required.
Why it matters
Preparing a system that embeds a protein in a lipid membrane typically requires careful alignment, estimation of packing densities, and avoiding atomic clashes. With Molecular Box Builder, this process is visual, interactive, and doesn’t require coding.
Step-by-step: From PDB to Bilayer
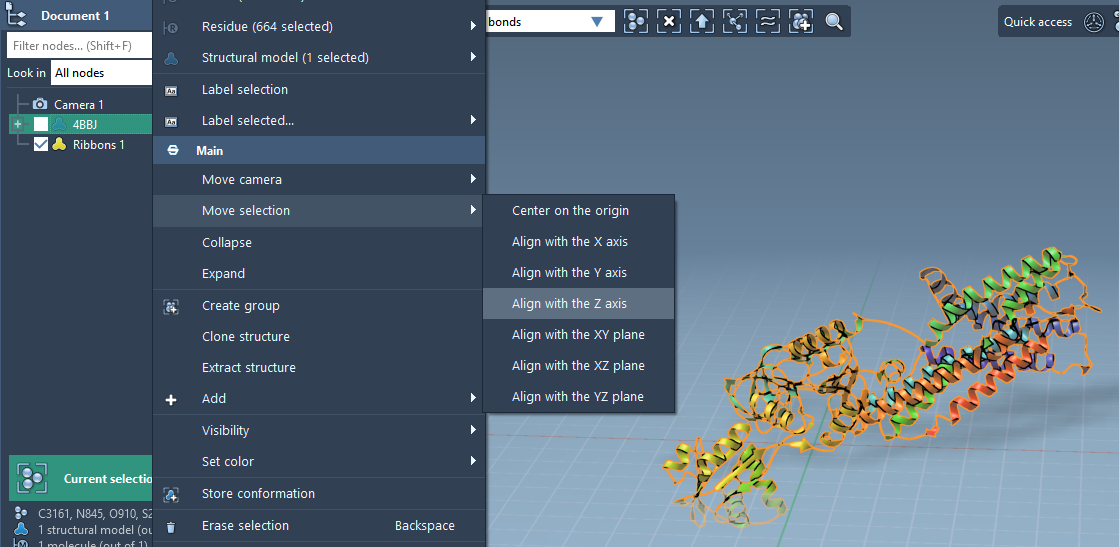
1. Align the protein
Start by importing your protein of interest, such as the copper-transporting PIB-ATPase 4BBJ. Right-click it in SAMSON’s Document View, then:
- Select Move selection > Align with Z axis to orient the protein vertically.
- Then select Move selection > Center on the origin to center it spatially.
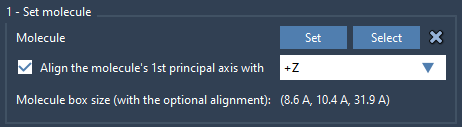
2. Add and align lipid molecule
Next, load a lipid molecule you’d like to use (e.g., POPC), select it in the Viewport or Document View, and click Set in the Molecular Box Builder app. Align its principal axis with +Z.
3. Define the box dimensions
In the app’s interface, define a box centered around the protein that fits a single lipid layer. You can tweak the margin between molecules to control spacing and avoid overlaps.
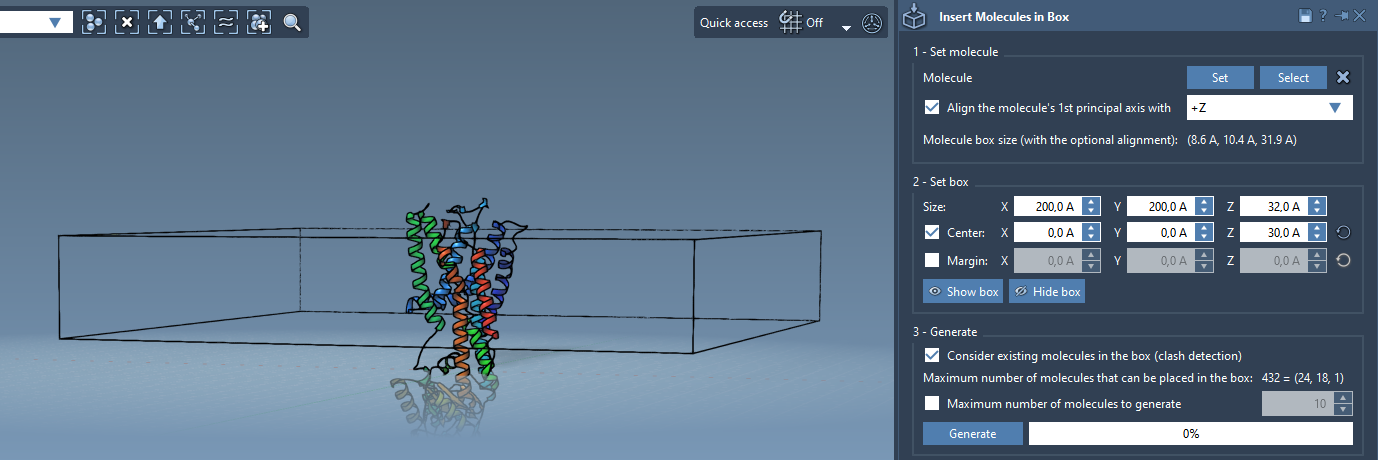
4. Generate the first lipid layer
Check Consider existing molecules in the box to avoid clashes with the protein, and click Generate. You now have a lipid monolayer embedding the bottom side of your protein.

5. Add the second leaflet (optional)
To form a full bilayer:
- Change the molecule’s alignment to
-Z. - Shift the box center slightly along the Z direction.
- Click Generate again to add the upper layer.
The result is a symmetric lipid bilayer system around your protein, created entirely within SAMSON’s visual environment — no exporting or scripting required.
What next?
After building your bilayer, you can minimize and simulate the system using the GROMACS Wizard directly from SAMSON. Or export the model to your preferred simulation setup.
Learn more in the full Molecular Box Builder documentation: https://documentation.samson-connect.net/tutorials/molecular-box-builder/molecular-box-builder/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





