One of the common tasks in molecular modeling is identifying which atoms or residues are close to one another. This is essential for analyzing interactions, binding pockets, or potential clashes. However, manually browsing molecular structures to find such proximities can be tedious and error-prone.
If you’re using SAMSON, proximity-based selection becomes significantly easier thanks to its Node Specification Language (NSL). This functionality lets you automatically select groups of atoms or residues that are within (or beyond) a specific distance of a structure or selection. This can be helpful in workflows like hydrogen bond detection, clash analysis, identifying nearby ligands, or visualizing residues around an active site.
Using Proximity Operators in NSL
SAMSON’s NSL allows proximity queries with intuitive keywords:
within [distance] of(short:w [distance] of)beyond [distance] of(short:b [distance] of)
Here’s a simple example. Suppose you want to find all carbon atoms within 5 angstroms of the residue named "GLN 2":
|
1 |
C w 5A of "GLN 2" |
Alternatively, if you’re interested in all atoms outside of a certain selection but still within a distance, NSL lets you combine proximity with logical composition. For example, to find atoms near but not in "GLN 2":
|
1 |
n.t a within 5A of "GLN 2" out of "GLN 2" |
Notice how detailed you can be with proximity searches. These expressions streamline your workflow by abstracting away the need for visual inspection or distance measurement tools. Whether you’re working with ligands, neighbors of a mutation site, or screening flexible docking poses, NSL’s proximity platform simplifies structural filtering to a single line.
Beware of Unexpected Matches
If your query seems to return more than expected — say you searched for anything within 5A of "GLN 2" and everything got selected — it might be because the query is matching higher-level nodes like folders or entire models that contain atoms within your range. To avoid this, bind your query to a more specific node type using n.t a (node type atom):
|
1 |
n.t a within 5A of "GLN 2" |
This ensures that only atoms, not containers, are selected.
Real-World Use Case
Let’s say you’re working with a protein-ligand complex and want to investigate the environment around the ligand. A practical NSL query would be:
|
1 |
n.t r w 4A of n.c lig |
This selects all residues (node type r) within 4 angstroms of any ligand (node category lig), letting you quickly identify potential binding interfaces.
Plus, you can use NSL directly within SAMSON’s Find tool or filter view in the Document view. For convenience, tapping Tab in the Find box offers auto-completion. SAMSON also integrates an AI Assistant to help you construct these queries based on your current structure hierarchy.
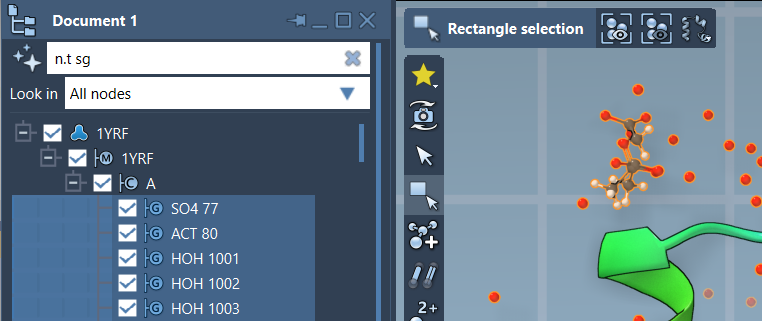
Once understood, NSL proximity queries become a fast and powerful way to navigate and interrogate molecular data.
To learn more about using proximity operators in SAMSON’s NSL, check out the official documentation: NSL Documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





