Designing molecular systems often involves experimenting with structures: forming bonds, breaking them, tweaking atom types. For many molecular modelers, these changes have traditionally required discrete steps such as redefining topology, restarting simulations, and recalculating parameters—interrupting the creative or scientific flow.
That’s where the Interactive Modeling Universal Force Field (IM-UFF) comes into play. Integrated into the SAMSON platform, IM-UFF goes beyond static molecular modeling by allowing smooth, real-time changes in molecular topologies.
What is IM-UFF?
IM-UFF builds on the standard Universal Force Field (UFF), extending it with support for interactive modeling. This means that instead of working with fixed topologies, you can manipulate atoms directly and see the effect on the system’s energy and structure—in real time. IM-UFF detects when bonds should form or break and automatically updates atom types and bond orders, all based on the positions of atoms.
This is invaluable when, for example, you’re exploring reaction mechanisms or assembling custom molecules. You can move atoms with the mouse and let the system adapt, reducing the need for manual re-typization or reinitialization.
How It Works
Once you’ve loaded a molecular system into SAMSON, all you need to do is:
- Add a simulator via Edit > Simulate > Add simulator.
- Select Interactive Modeling Universal Force Field as your interaction model.
- Choose a state updater (e.g., FIRE).
- Uncheck the Static topology (UFF only) option to enable topological changes.
- Start the simulation and begin manipulating atoms.
During the simulation, you can:
- Break bonds by pulling atoms apart.
- Form new bonds by bringing atoms close together.
- Modify atom types and connectivities automatically based on current structure.
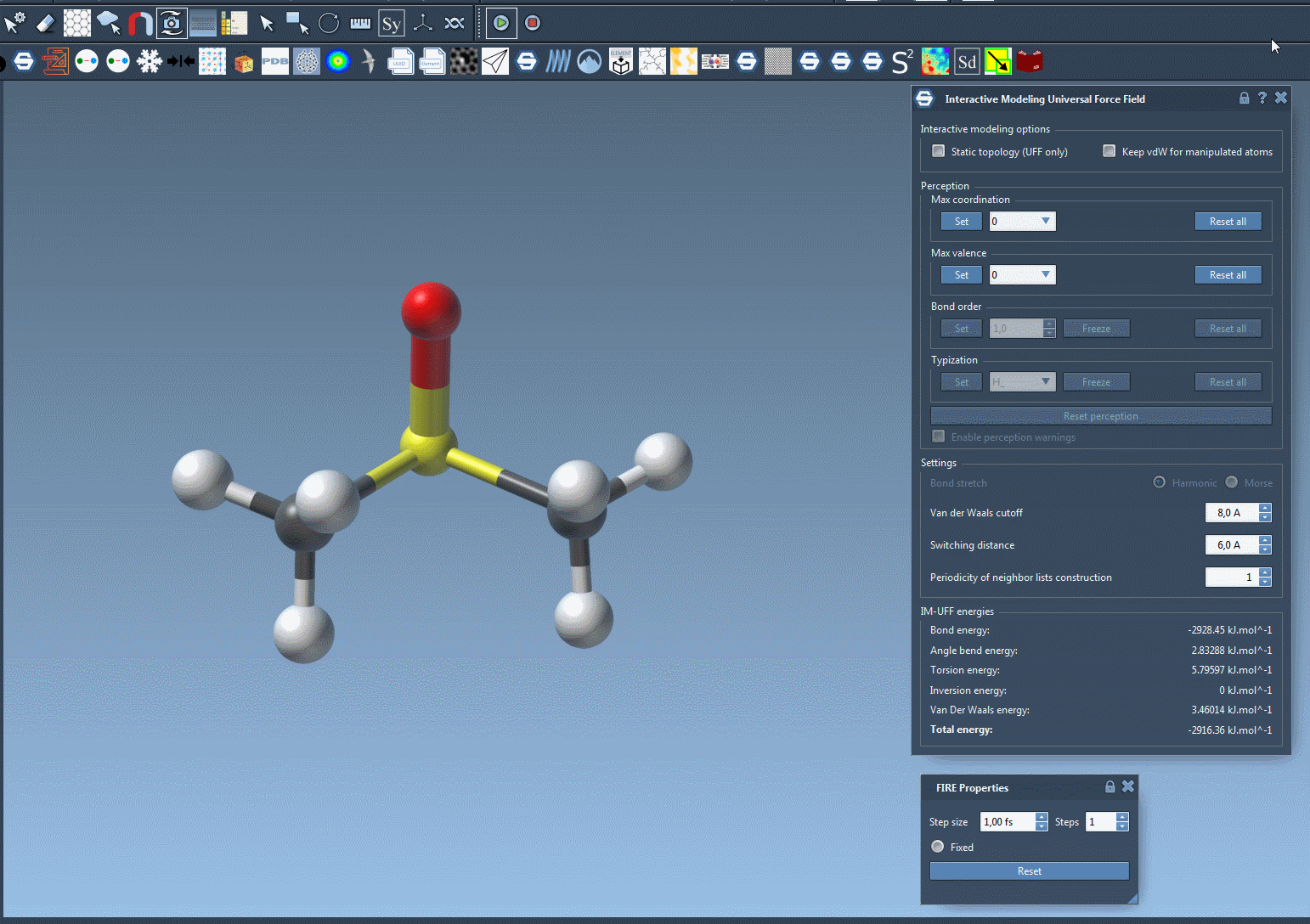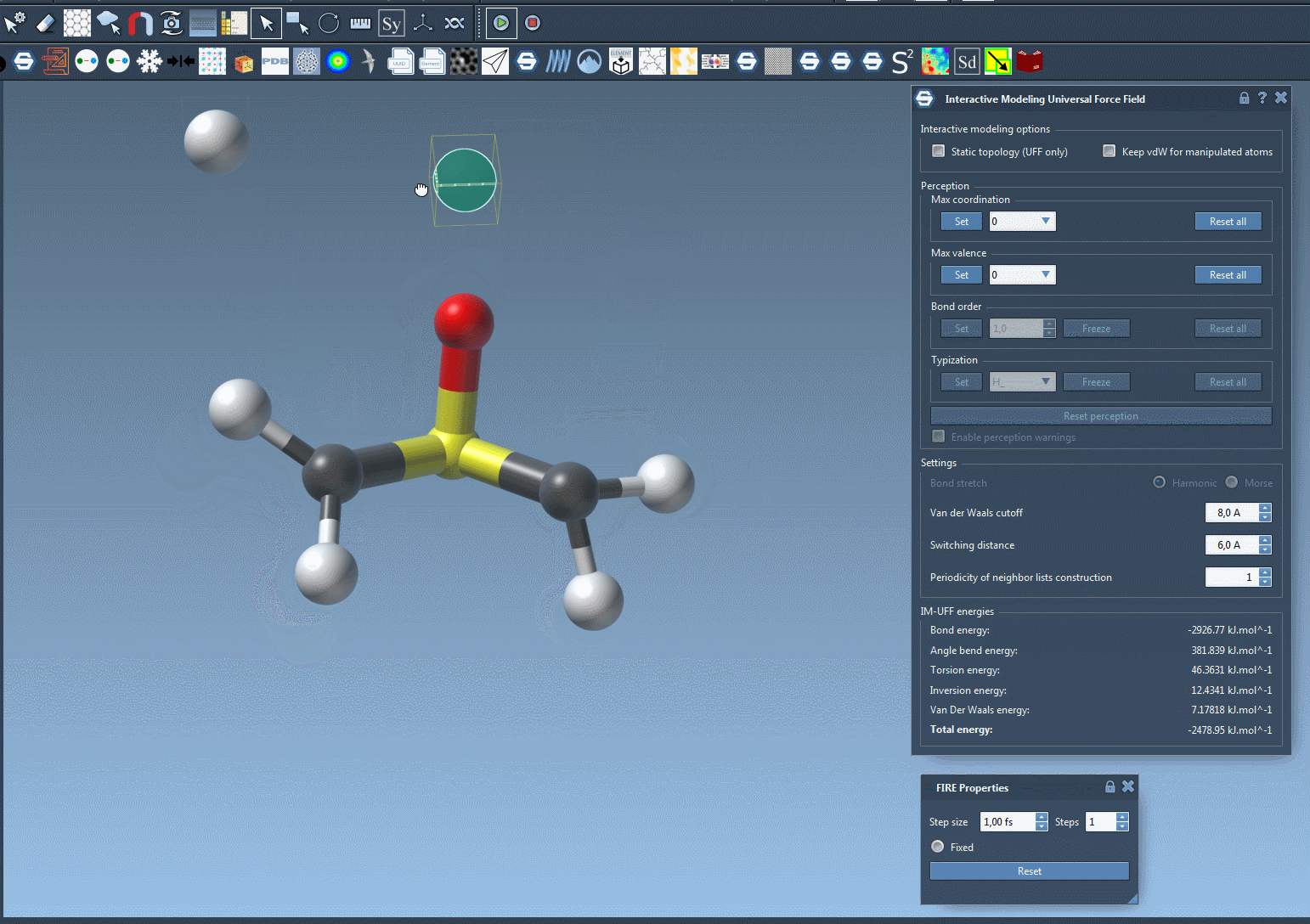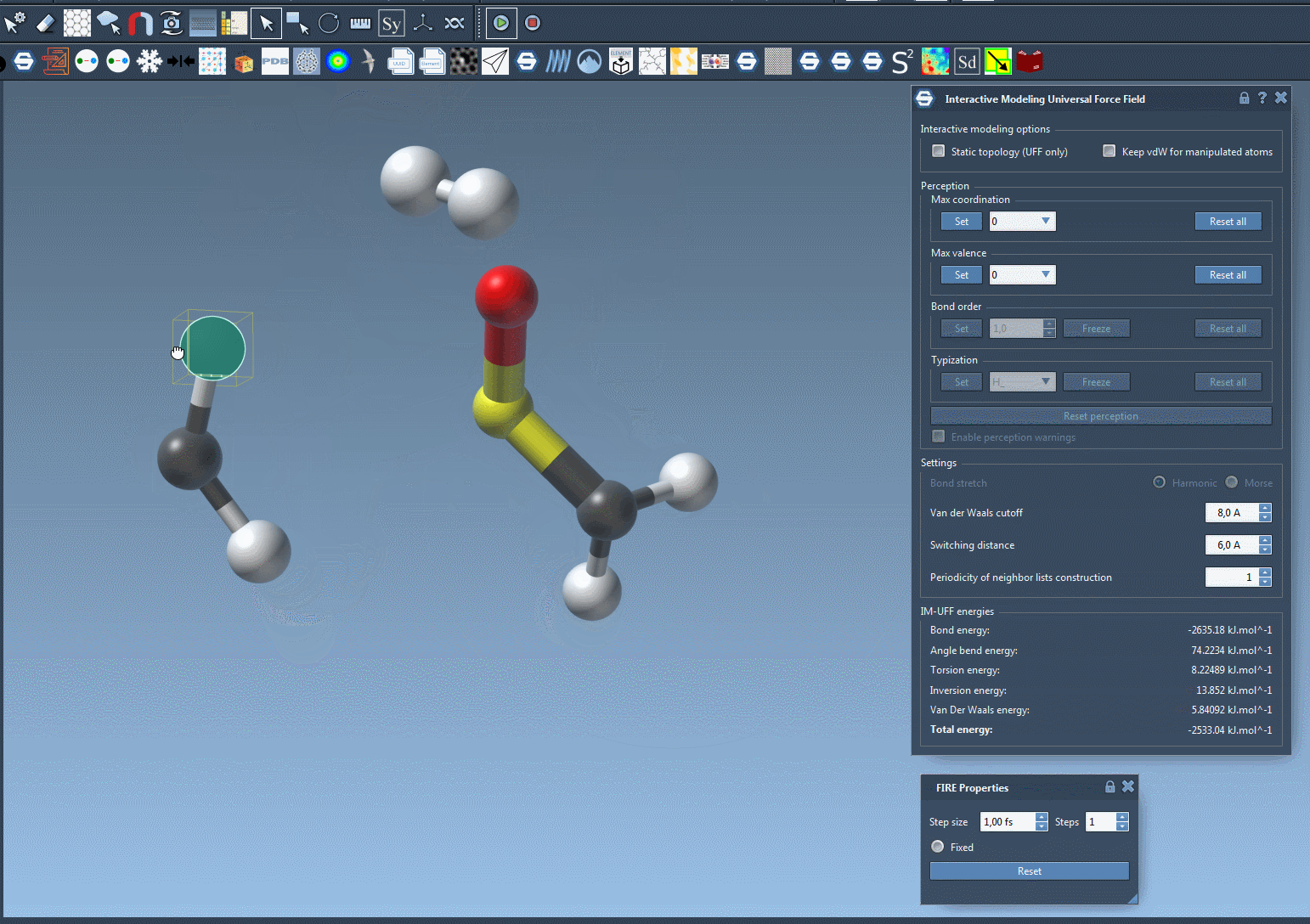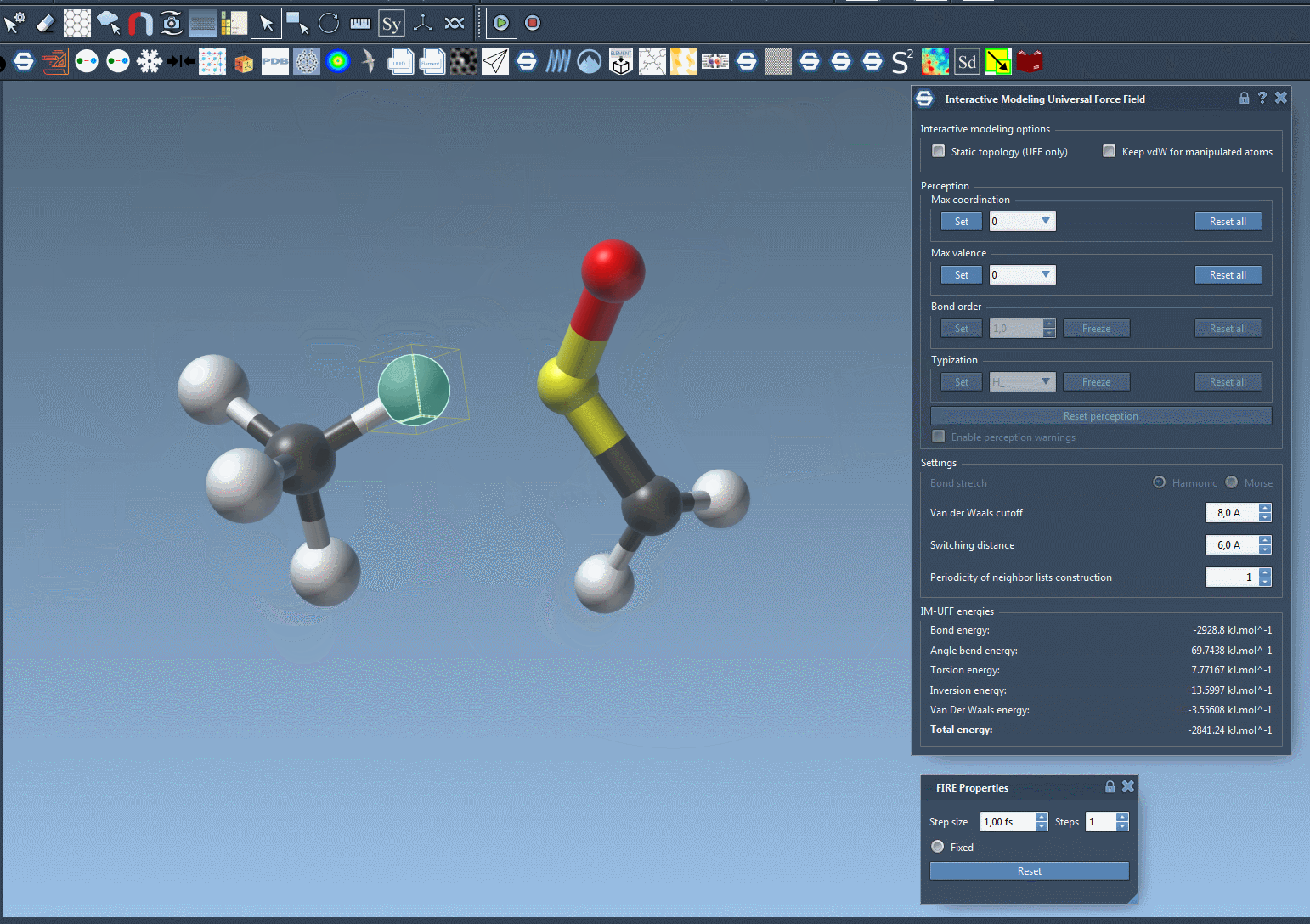
Why This Matters
Being able to interactively edit topology brings flexibility into workflows such as:
- Simulating chemical reactions without explicitly encoding bond-breaking or forming steps.
- Building new molecular architectures step-by-step with physical guidance.
- Creating molecular animations or educational demos that reflect realistic dynamics.
Plus, having access to continuously updated energy metrics helps you understand whether a structure is feasible or not—without needing to rerun a simulation every time you tweak something.
Best Practices
If you want smoother manipulations, you can disable van der Waals (vdW) interactions for manipulated atoms using the option Keep vdW for manipulated. This is helpful when positioning atoms for bonding, especially since vdW repulsion might prevent close approach. Simply uncheck this setting to make manual adjustments easier.
IM-UFF isn’t just useful for researchers—students and educators can also benefit from this exploratory, hands-on approach to building molecular systems.
Want to Try It?
You can start using IM-UFF by installing it from within SAMSON and loading your molecular models. For a more detailed guide, including how to customize simulation parameters, check the official documentation:
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





