One of the challenges many molecular modelers face is the ability to predict biomolecular structures that go beyond plain protein sequences. If you’re a researcher working with protein-ligand systems, modified nucleotides, or hybrid biomolecules, you’ve likely encountered limitations in traditional prediction tools.
With the Boltz-2 prediction service in the Biomolecular Structure Prediction extension in SAMSON, you can now include ligands and sequence modifications directly into your prediction workflows.
What Makes Boltz-2 Different?
Most structure prediction methods focus on obtaining 3D structures from amino acid or nucleotide sequences. However, real-world systems often include small molecule ligands or modified residues that play essential biological roles. Boltz-2 enables you to:
- Add proteins, DNA, and RNA sequences.
- Include ligands using SMILES strings or CCD codes.
- Introduce modifications at specific residues using CCD codes.
Step-by-Step: Predicting with Ligands and Modifications
Here’s how to use Boltz-2 in SAMSON for accessible, more detailed structure prediction:
- In SAMSON, go to Home > Predict.
- Choose the Boltz-2 service.
- Use the Add protein, Add DNA, Add RNA, or Add ligand buttons to define your system. You can input single or multiple types as needed.
- To add a chemical modification, click Add modification. You’ll be prompted to select the modified residue’s position and enter its CCD code. This is useful for modeling methylations, fluorescent labels, and other non-standard elements.
- Once your components are configured, click Start prediction. Calculations are done in the cloud using high-performance A100 GPUs.
Optimize Your Predictions
Each prediction with Boltz-2 uses computing credits, typically ranging between 0.5–1 credit per structure. This allows you to scale your prediction workload based on your needs. If you’re working on multiple designs or iterative modeling, having the flexibility to selectively include detailed components like ligands or base modifications can save significant time in downstream analysis.
You can monitor the progress of your predictions from the Interface > Cloud Jobs section within SAMSON or on the Jobs page on SAMSON Connect.
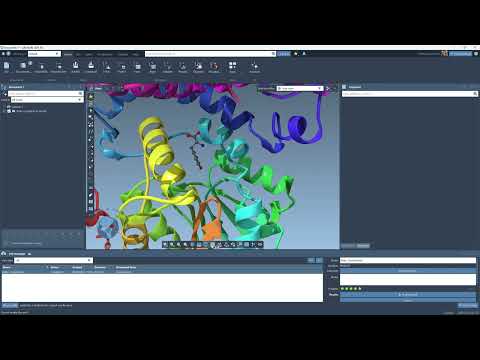
Learn by Watching
To get started quickly, check out this short video tutorial to see how to run simulations using Boltz-2 and generate protein-ligand interaction diagrams—all in under five minutes.
Why This Matters
Being able to model modified structures and liganded complexes from the start helps avoid the need to manually patch structures later, which is often error-prone and time-consuming. Boltz-2 simplifies this process by integrating it into the prediction workflow in SAMSON.
To learn more about the Biomolecular Structure Prediction extension, including Boltz-2 and other services, check out the full documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





