Generating lipid bilayers around proteins is a common step in membrane protein modeling, but it often involves multiple tools, file format conversions, and error-prone setup steps. If you’ve ever found yourself frustrated by having to script this process—or worse, trying to get molecules to align manually—this guide is for you.
SAMSON’s Molecular Box Builder extension makes constructing lipid layers and bilayers around proteins a visual, guided, and user-friendly process. Here’s how to do it step-by-step, directly in the SAMSON platform.
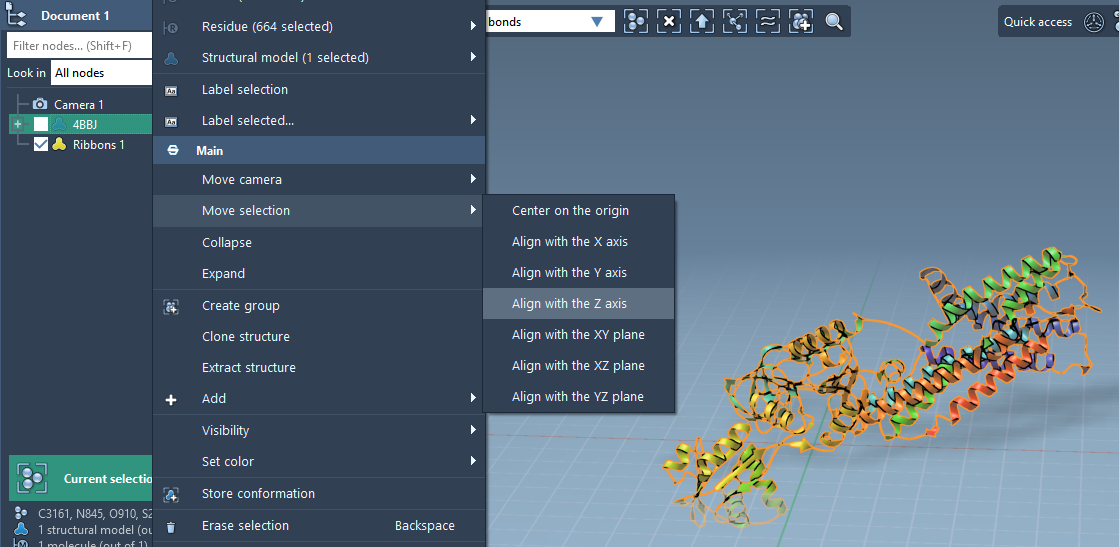
Aligning the Protein
Most membrane proteins should be oriented such that their transmembrane axis aligns with the Z axis. In SAMSON, this is a simple action:
- Find your protein in the Document view.
- Right-click and choose Move selection > Align with Z axis.
- Then center it using Move selection > Center on the origin.
This alignment ensures the lipid bilayer will form symmetrically around the embedded protein.
Setting the Lipid Molecule
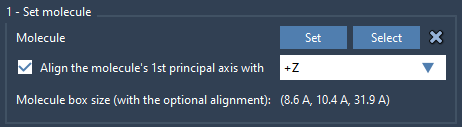
You can import a lipid structure from your own database or from online resources. Once loaded in SAMSON:
- Select the lipid molecule in the Document view or Viewport.
- Open the Molecular Box Builder and click Set.
- Align its principal axis to the
+Zaxis in the app.
Defining the Box
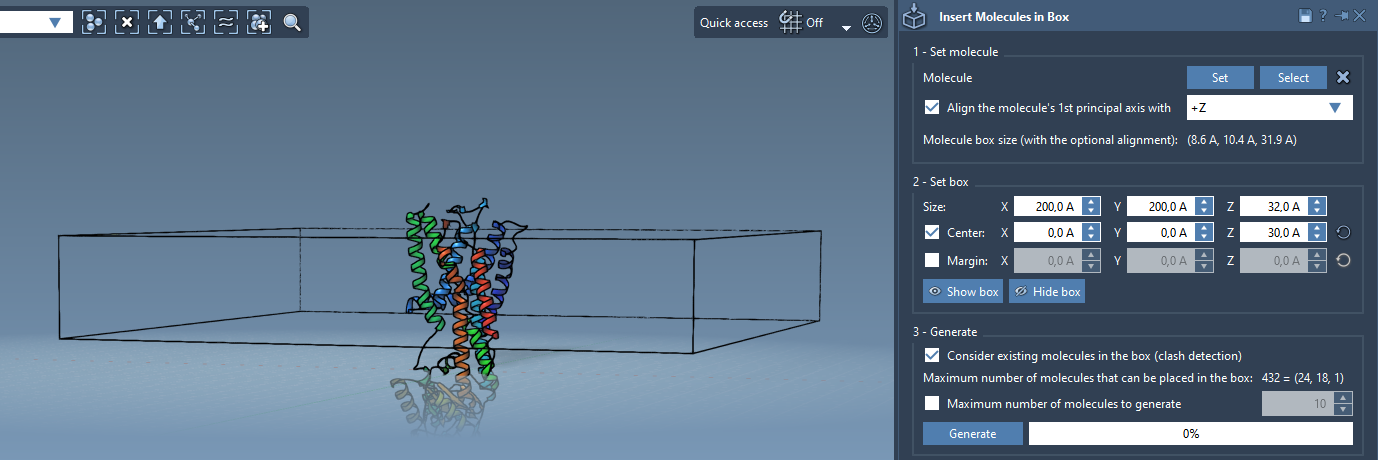
The idea here is to define a 3D box that envelopes the protein and has dimensions suitable for a membrane layer. You can:
- Center the box around the protein.
- Adjust the X and Y dimensions to extend slightly beyond the protein’s lateral size.
- Use the Z dimension to define how thick your membrane layer will be.
Generating the Lipid Layer
You can now choose to populate the box with lipid molecules:
- In the app, enable Consider existing molecules in the box. This prevents overlap with the protein.
- Click Generate—the app fills the layer with lipids placed around the protein.

Optional: Creating a Bilayer
If you want a true bilayer:
- Set up and generate the first layer using alignment to
+Z. - Shift the box center downward along Z (e.g., by the thickness of the membrane).
- Change the lipid orientation to
-Zfor the second layer and generate again.
This approach gives you a controlled and reproducible way to set up bilayers quickly—no scripting or manual manipulation required.
To explore more advanced features or to follow the complete guide, visit the official Molecular Box Builder documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





