Molecular modelers working with large symmetric assemblies—like viral capsids or multi-subunit protein complexes—often face a dilemma: Should I simulate the entire structure, or is there a smarter way?
In many cases, the repetitive nature of these biological assemblies means you’re simulating the same thing over and over. That’s expensive and inefficient. If you’ve ever waited hours for a simulation to finish, only to realize you could have focused on a smaller representative part, this post is for you.
What Is an Asymmetric Unit?
An asymmetric unit is the smallest portion of a molecular structure that can be used to reconstruct the whole assembly through symmetry operations. It contains all the unique information, without redundancy.
Simulating just the asymmetric unit—when symmetry allows—can drastically reduce computational cost, without losing accuracy in many applications.
How to Identify and Extract an Asymmetric Unit with SAMSON
SAMSON’s Symmetry Detection extension makes it easier to identify the unique portion of a structure. Here’s how:
- Open SAMSON and load a biological assembly (e.g., the PDB structure
3NQ4for an icosahedral virus). - Launch the symmetry detection tool: Home > Apps > Biology > Symmetry Detection.
- Click Compute symmetry to find all axes and possible symmetry groups.
- Explore identified symmetry axes and groups. For large structures like
3NQ4, SAMSON detects all 2-, 3-, and 5-fold symmetry elements typical of icosahedral symmetry. - Choose a symmetry group with low RMSD (to ensure accurate repetition across elements).
- Identify and highlight the asymmetric unit, and export it as a new model ready for simulation.
This workflow is especially powerful for cases where visualizing the entire assembly isn’t feasible due to hardware limits, or when minimizing computational resources is critical (such as during coarse-grained modeling, force field tuning, or early-stage exploratory simulations).
Example: The 3NQ4 Icosahedral Capsid
When symmetry is computed for the large viral capsid 3NQ4, SAMSON displays its full icosahedral symmetry. Each of the axes (2-fold, 3-fold, 5-fold) is shown visually, aiding users in identifying the smallest repeat unit.
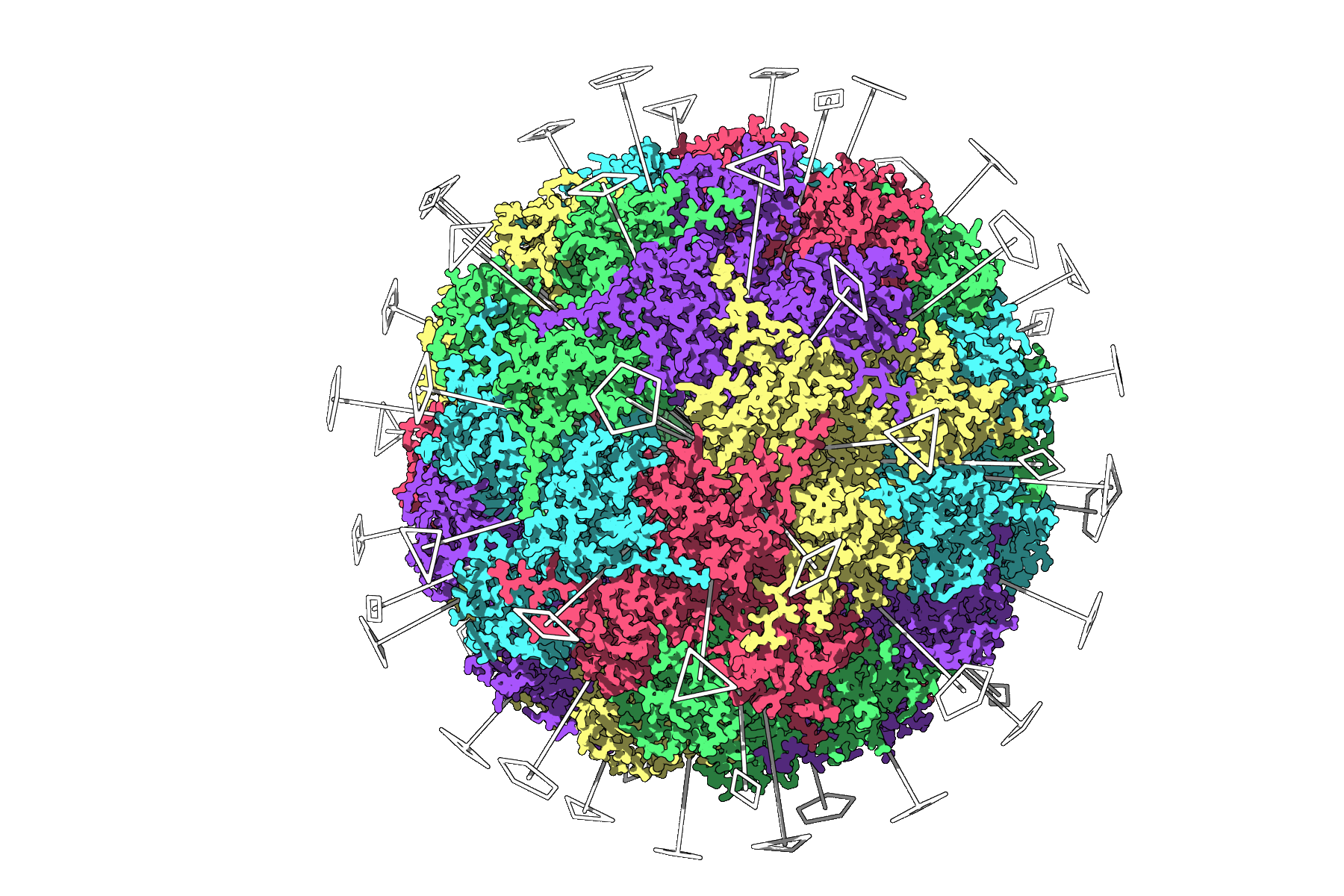
Tips for Better Visualization
- Use ribbon or surface views to clarify how units repeat spatially.
- Color individual chains or domains differently to quickly spot symmetry-related copies.
- Use Viewport snapshots to share the results or include visuals in your research documentation.
Time-saving tip: You can double-click an axis in SAMSON to reorient the camera and view symmetry head-on, which is useful for identifying planar or radial repetition.
Why This Matters
Cutting down a simulation from millions of atoms to just the unique unit avoids redundant calculations. This can be the difference between getting a result overnight or waiting days. It’s also essential when iterating over design changes—like mutating parts of symmetrical nanoparticles or tuning binding interfaces on symmetric oligomers.
To learn more about symmetry detection and working with asymmetric units in SAMSON, check out the full documentation: Symmetry Detection in Biological Assemblies.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON from https://www.samson-connect.net.





