When setting up calculations for reaction coordinates or preparing data for free energy profiling, researchers often find themselves in need of atomic coordinate files at multiple points along a ligand’s movement path — usually in PDB format. Doing this manually can quickly become repetitive and error-prone, especially when dealing with hundreds of frames.
The Export Along Paths extension in SAMSON offers a targeted solution to this common pain: it allows molecular modelers to automatically export the coordinates of only the atoms that matter — such as a ligand — along pre-defined paths.
Why export a subset?
In many workflows, such as enhanced sampling simulations, the focus isn’t on the full system, but rather on a part of it — like the ligand or active site residues. Exporting only this subset can significantly reduce post-processing time while improving clarity.
Here’s how to do it in SAMSON
Assuming you’ve already generated an unbinding path using, for example, the Ligand Path Finder extension, and have loaded your system into SAMSON, follow these steps to export only the ligand atoms along the trajectory path:
- Open the Export Along Paths app from Home > Apps > All > Export Along Paths.
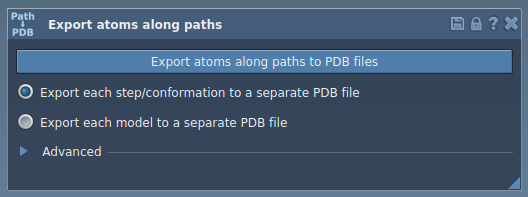
- In the GUI, expand the Advanced panel.
- Select your ligand (e.g.
TDG) in the Document view.

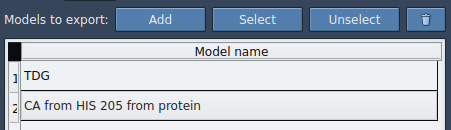
- Click Add in the app interface to define this selection as a model to export.

- Optionally, rename the model and add others if needed. When ready, select the path(s) in the Document view and choose your export format (one file or one per frame).
- Click Export atoms along paths to PDB files, choose a destination folder and file prefix, and you’re done.
What you get
- PDB files containing atomic coordinates of only the specified atoms (e.g., ligand) as they move along the path.
- Option to export all frames in a single file or each frame in its own file.
- Custom naming and grouping of multiple atom subsets for multi-ligand or focused site analysis.
Common use cases
- Generating structures for umbrella sampling calculations.
- Preparing snapshots for visual inspection or publication.
- Isolating key atoms (like a ligand) for use in downstream simulation packages.
This reduces complexity in post-processing and enables researchers to streamline their trajectory-related tasks from within an interactive interface.
To learn more, visit the official documentation page here.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





