When designing new compounds, one common goal is to understand how small structural modifications impact molecular properties. For medicinal and computational chemists, this often means repeating tedious edits, structure generation, and validation steps by hand. What if you could automate parts of this process, and visualize the results directly as they are generated?
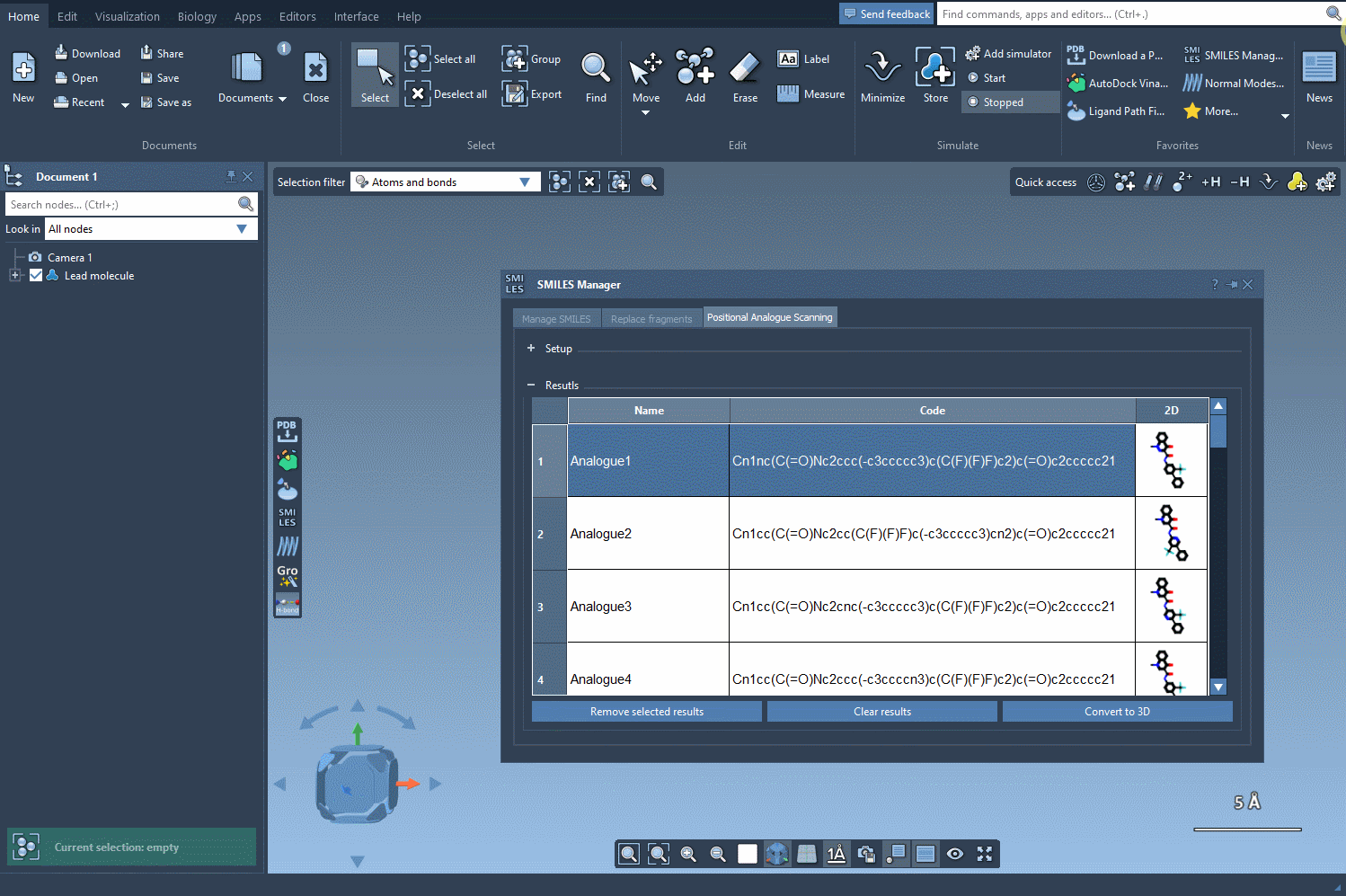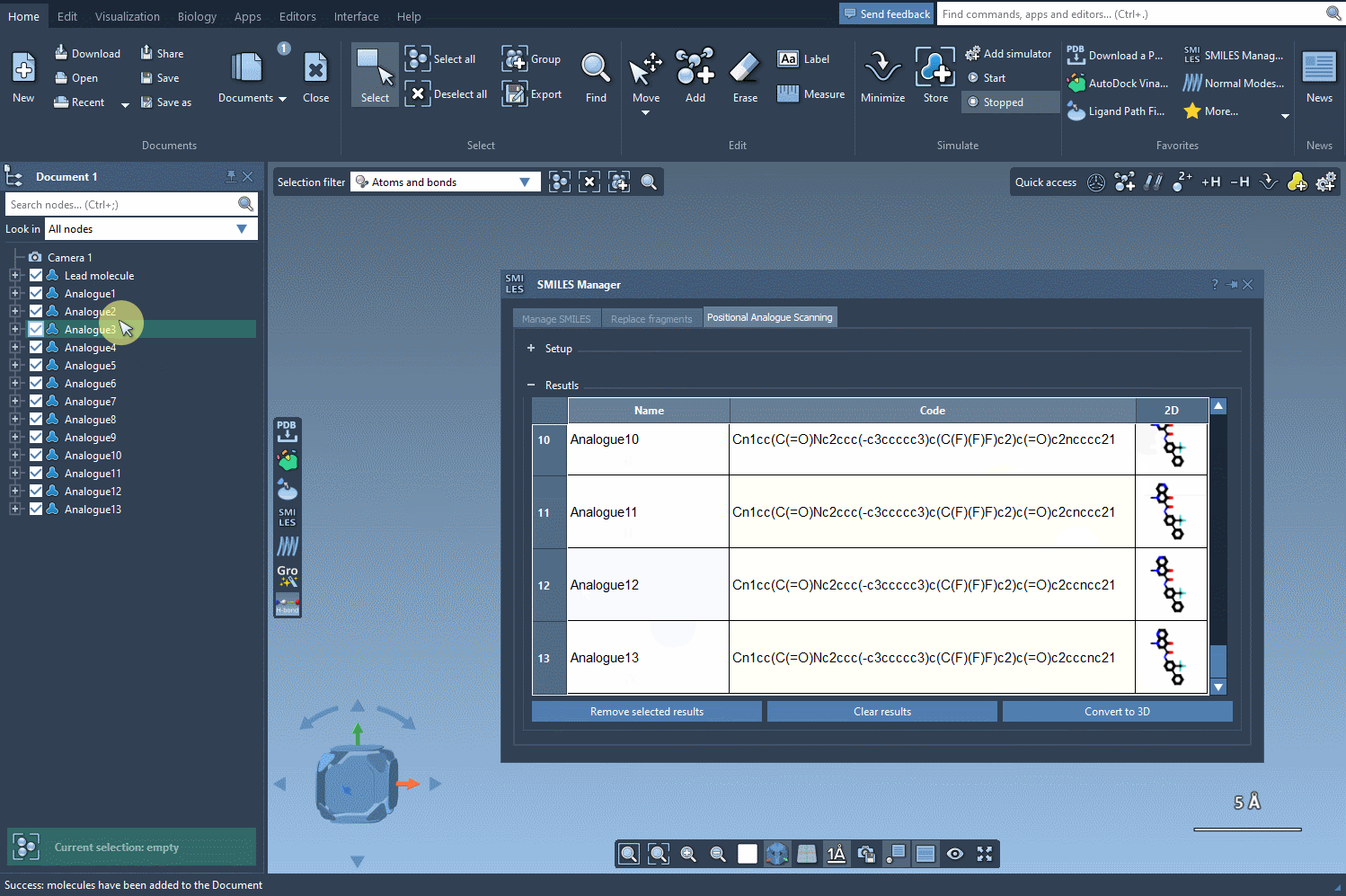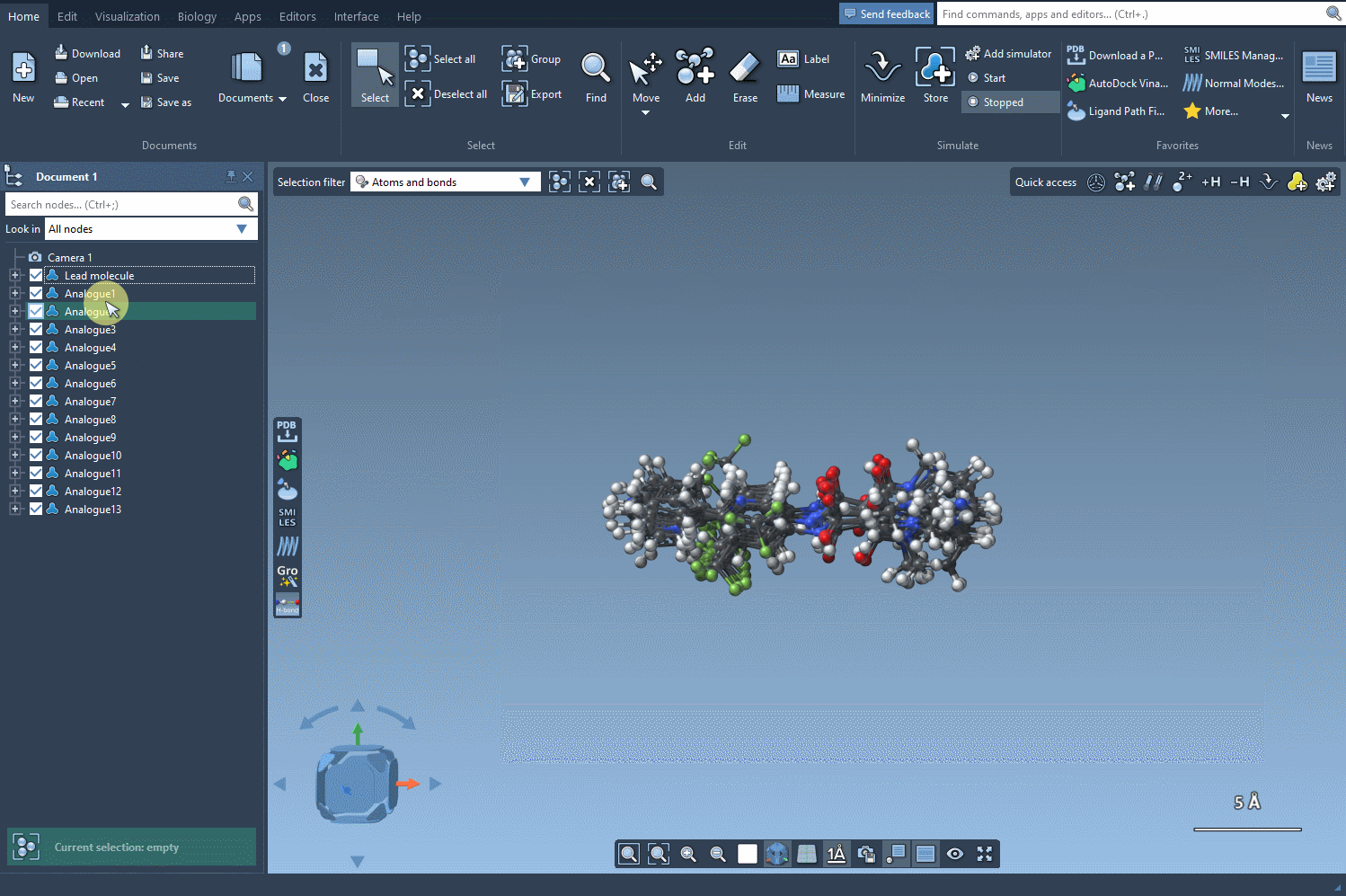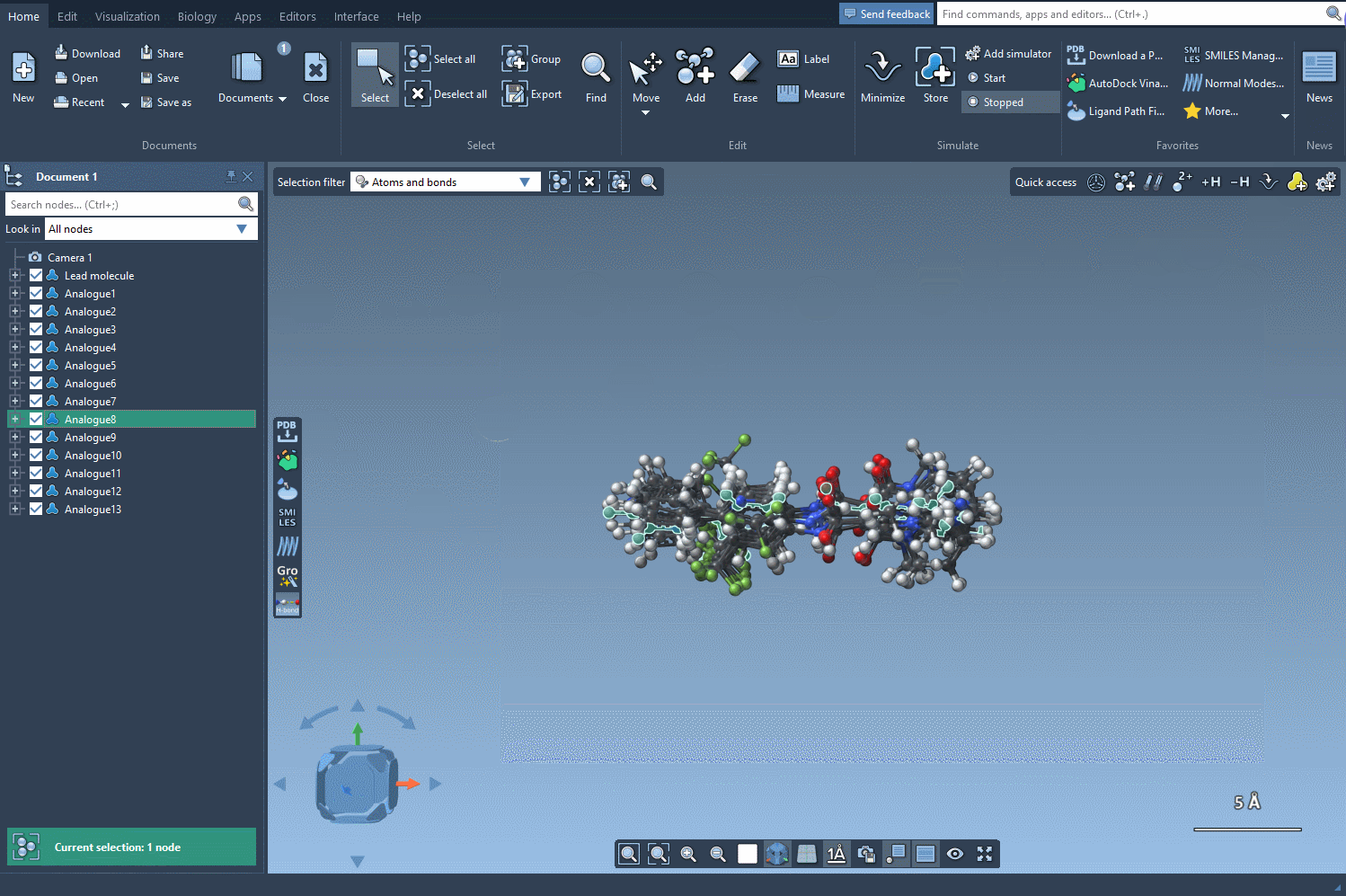
The SMILES Manager extension in SAMSON allows you to perform positional analogue scanning—a technique that systematically replaces or modifies specific atomic patterns in a molecule with alternative atoms or groups. In a few clicks, you can generate dozens of analogs from a single starting molecule, visualize their 2D structures, tweak individual analogs, and even generate 3D conformations—all within the same interface.
Why Positional Analogue Scanning Matters
Understanding small changes in molecular structure can provide insight into binding affinity, stability, and specificity. For example, changing a hydrogen atom to a fluorine, or a carbon to a nitrogen, may unlock better interactions with a target protein. But doing this manually, especially on large datasets or complex molecules, is not scalable.
Start with a Molecule
To begin, load or select your molecule in SAMSON. The extension lets you import molecules using SMILES strings or select structures already in your project. In this example, a known compound with SMILES: CN1C=C(C(=O)Nc2ccc(-c3ccccc3)c(c2)C(F)(F)F)C(=O)c2ccccc12 is used as the starting point.
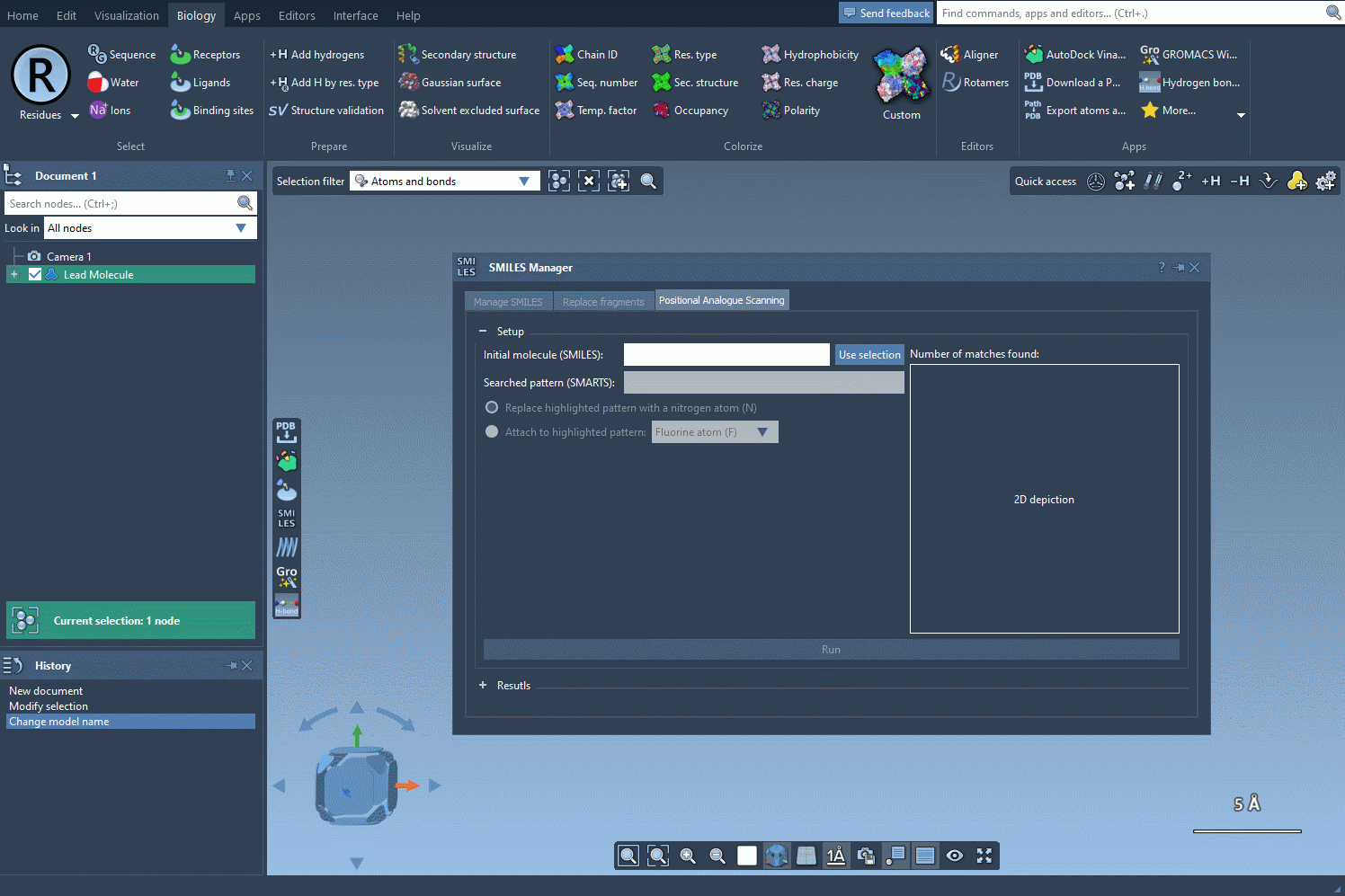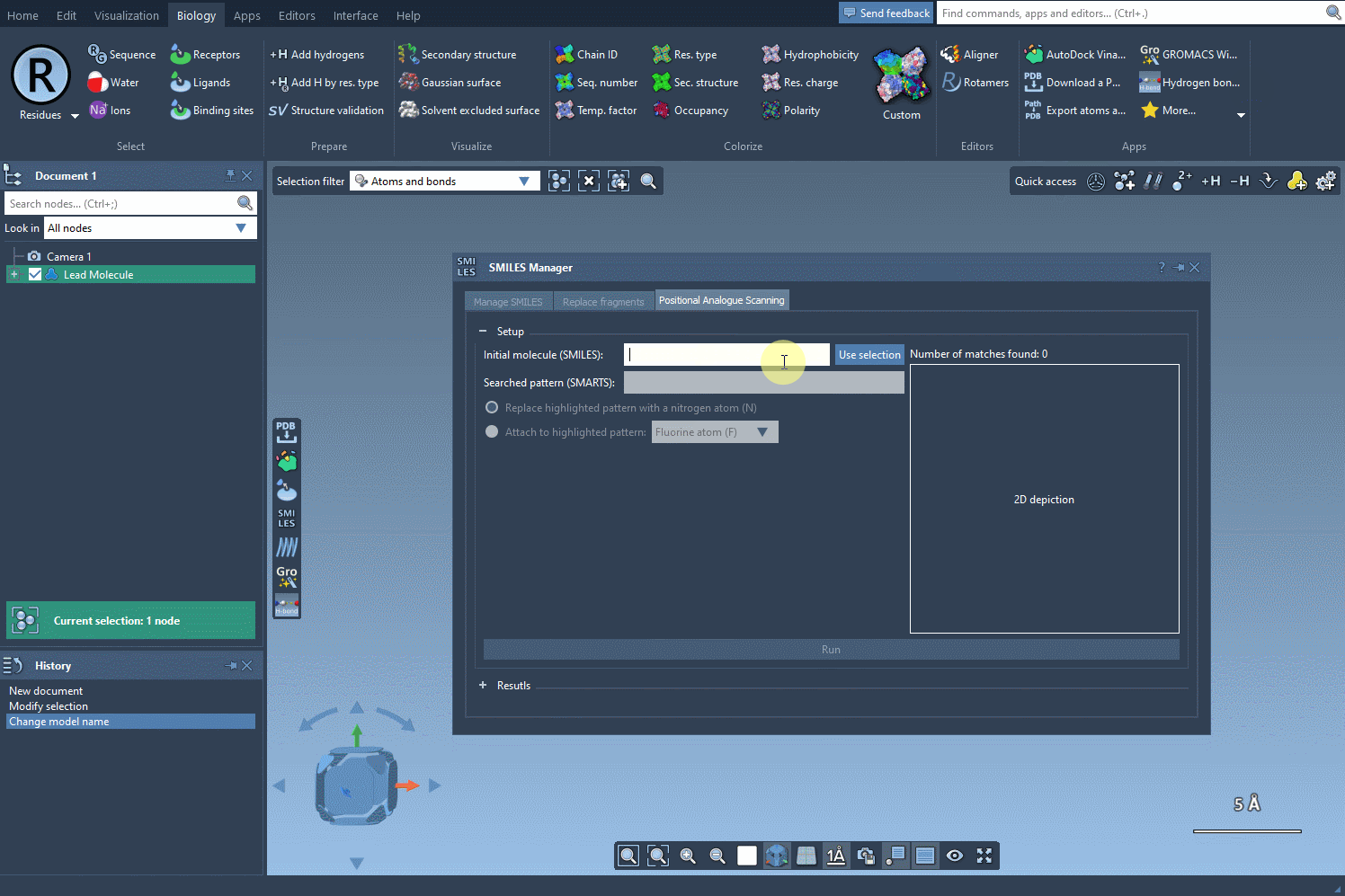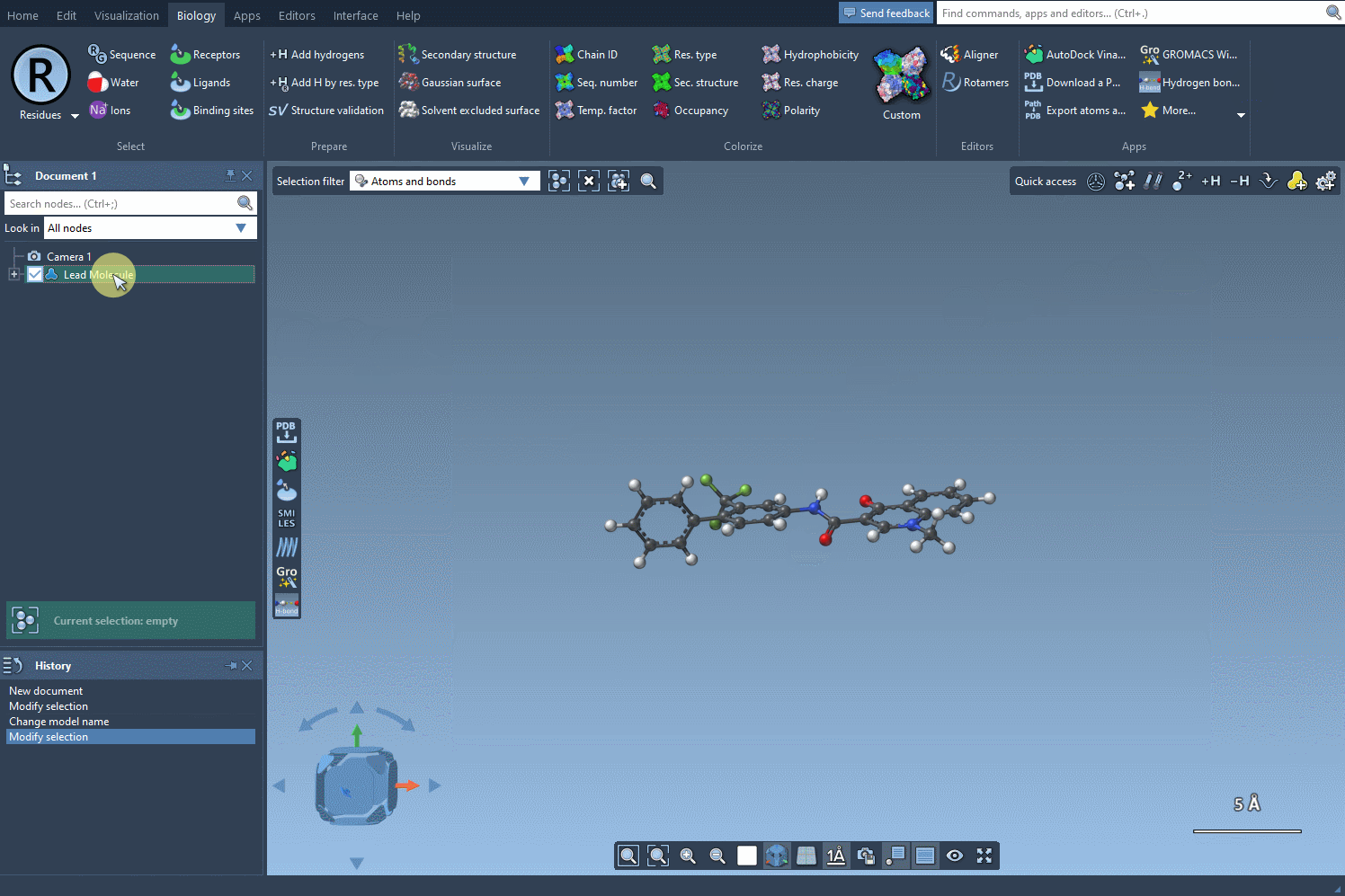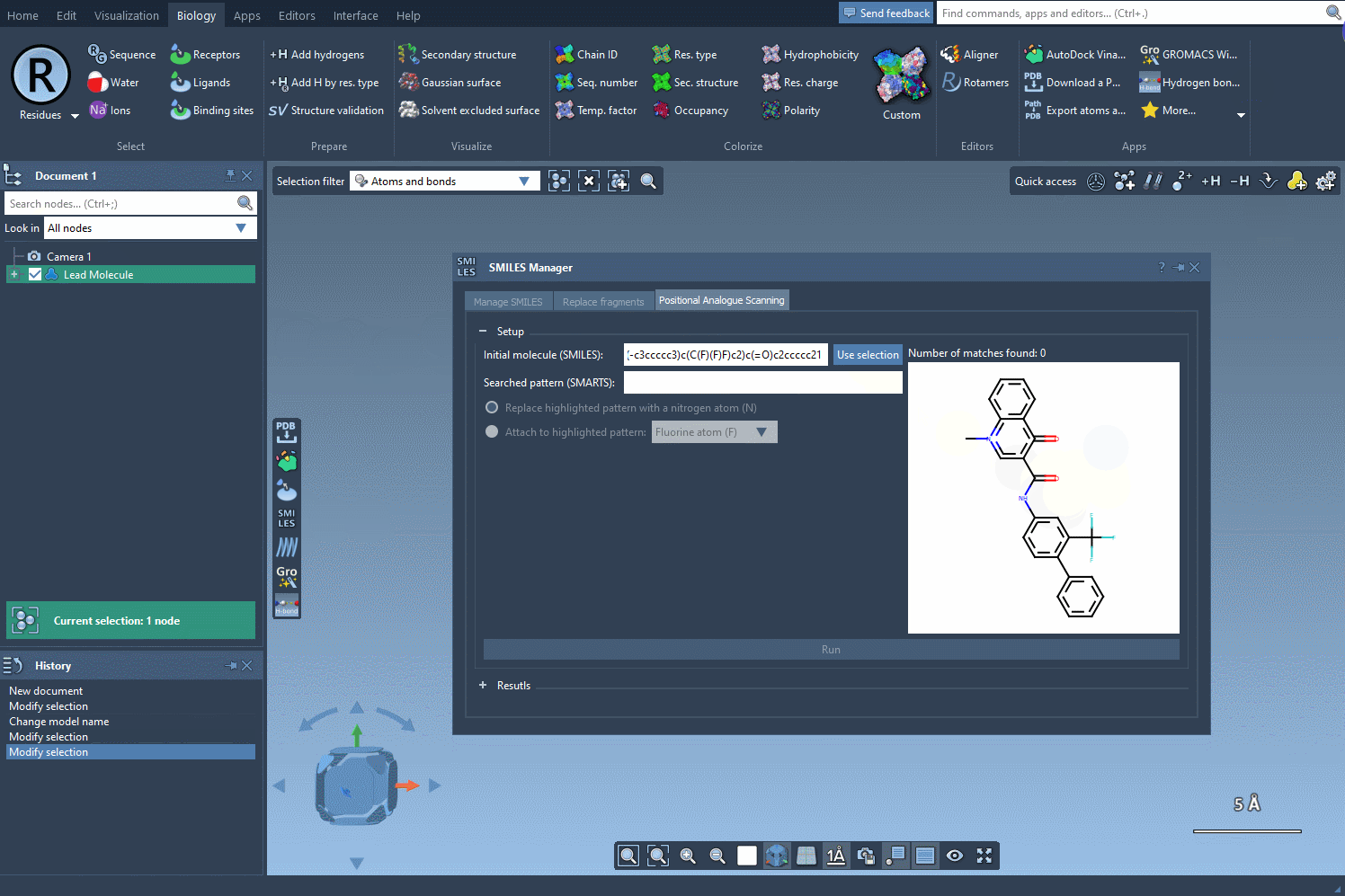
Define the Pattern You Want to Modify
Use SMARTS patterns to identify parts of your molecule to target. For instance, entering [cH] will match aromatic hydrogens in the structure. When applied, the matched atoms are highlighted so you can easily verify hit sites.

Modify and Generate Analogues
Next, choose how to alter the pattern. You can replace it (e.g., an aromatic C-H with N), or choose to attach new groups (like F or CH3). Then, hit Run and watch how the analogs are generated along with their 2D depictions.
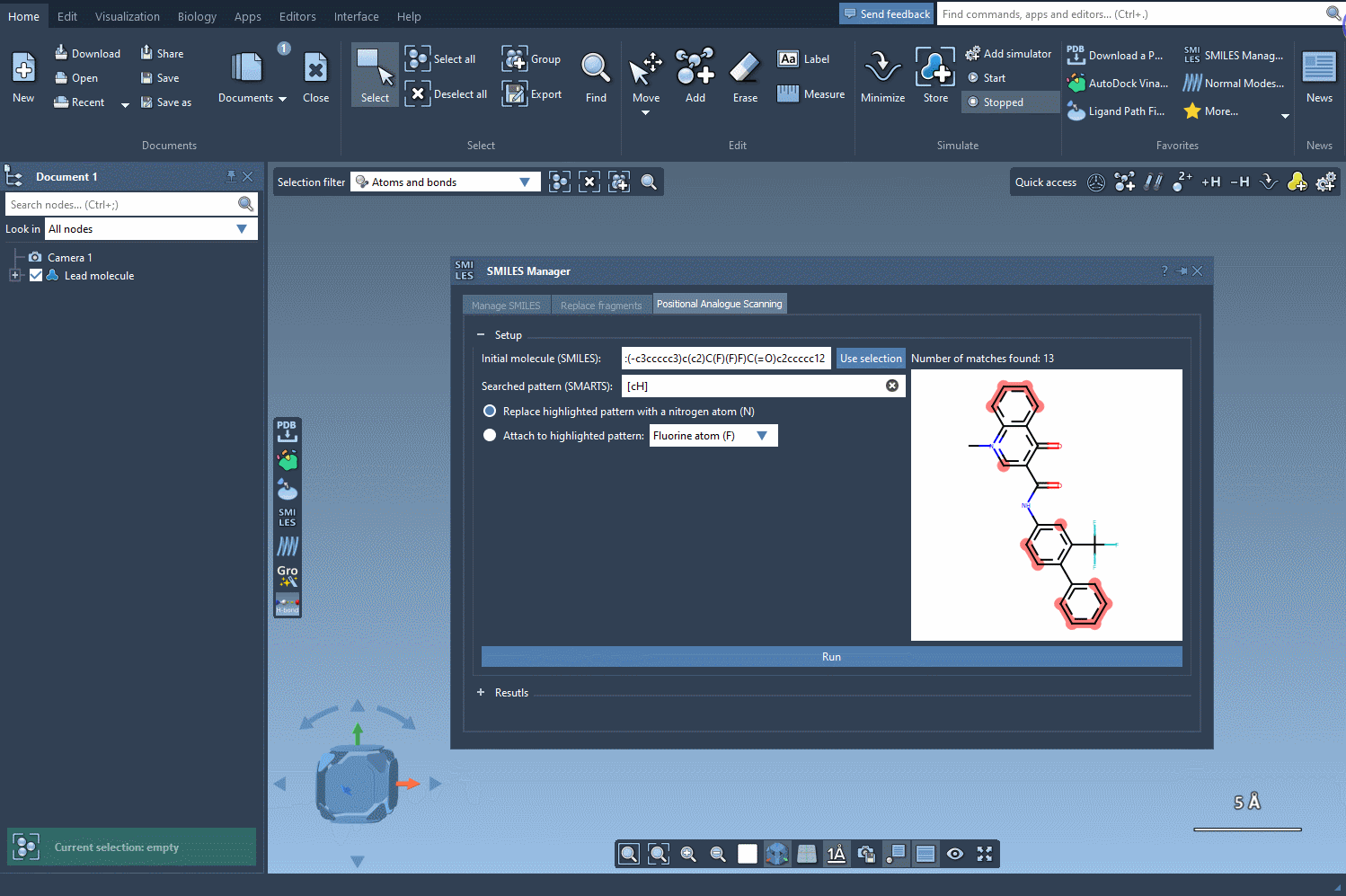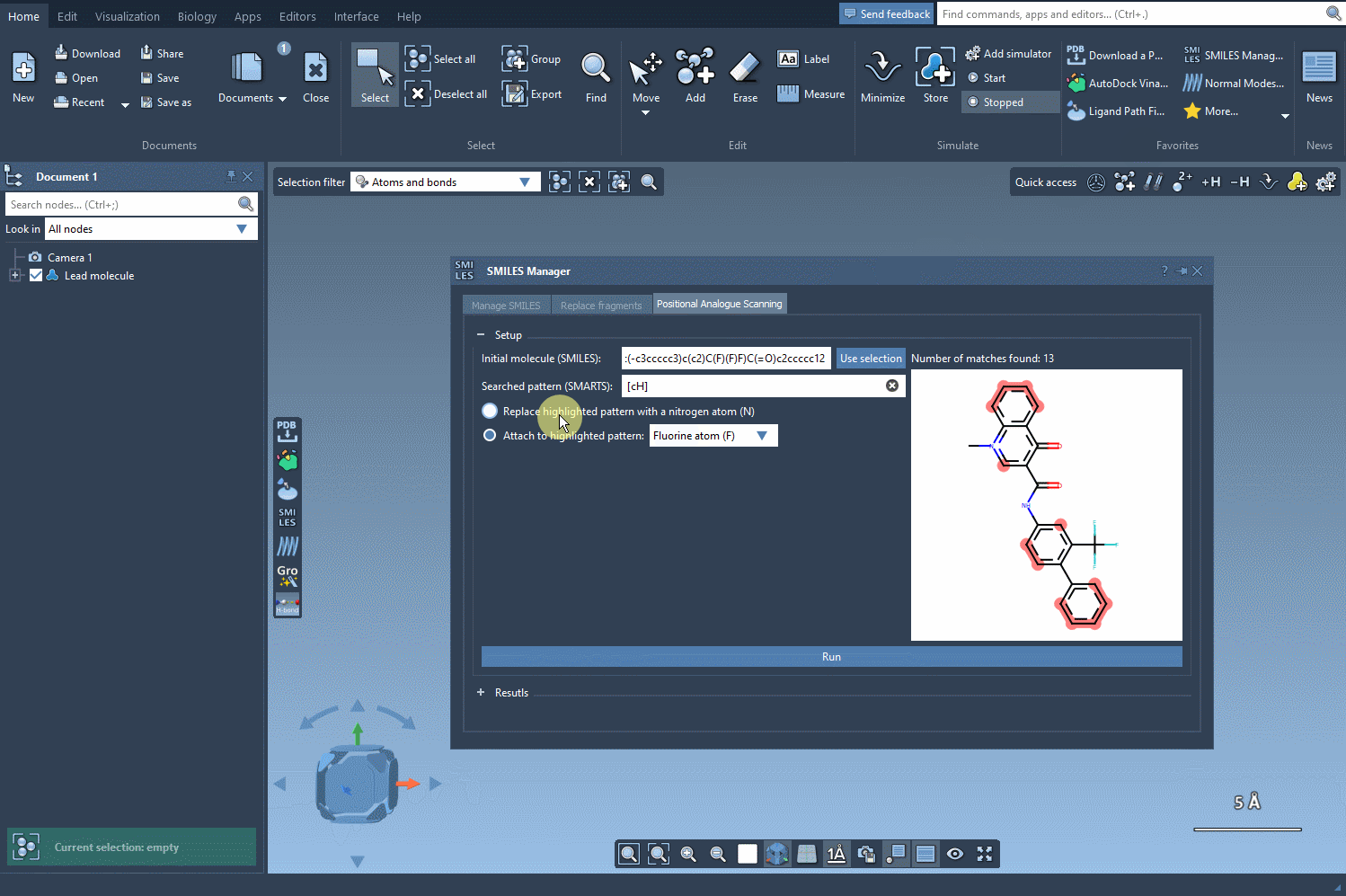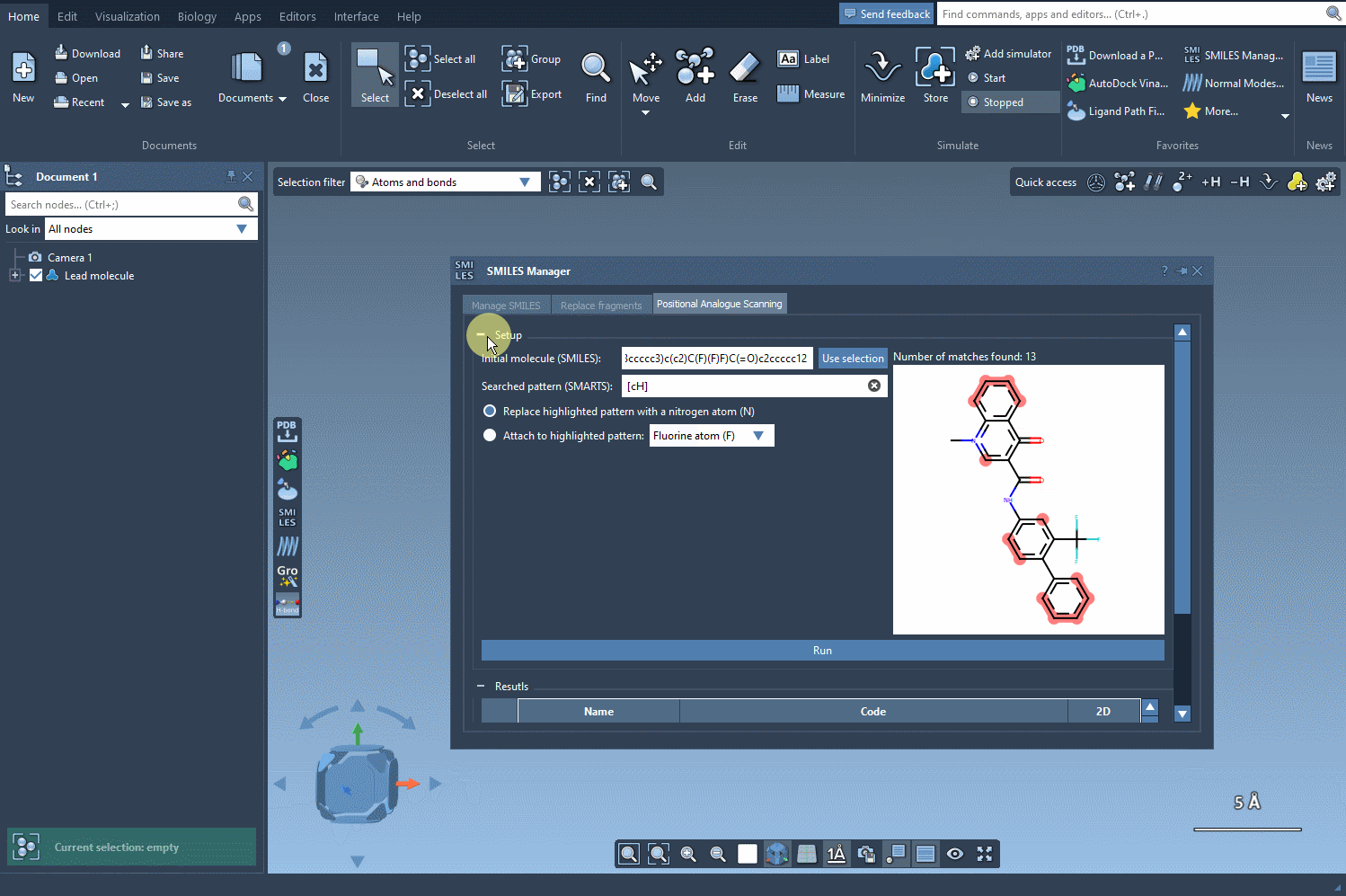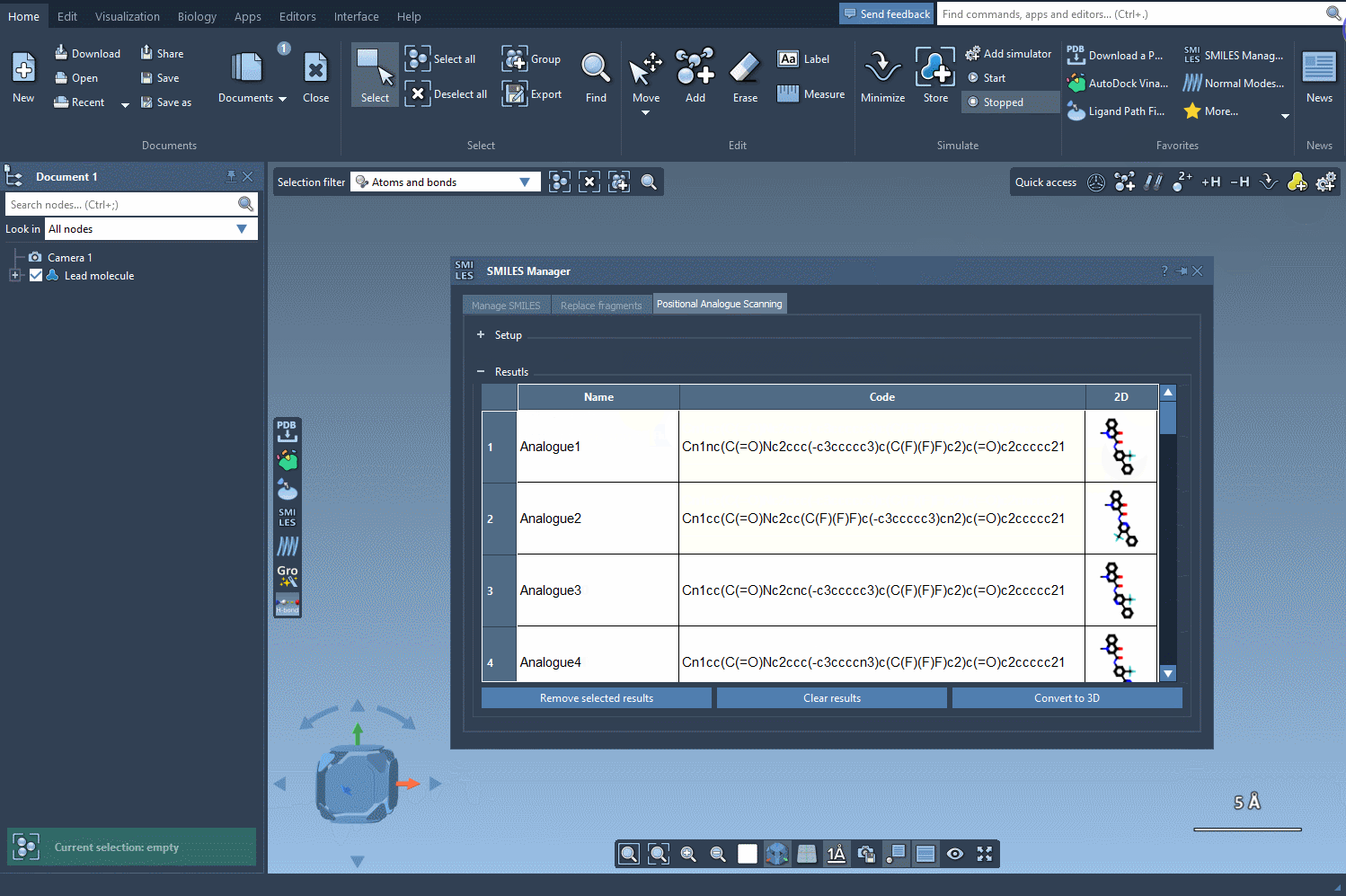
Visualize and Tweak Results
The generated analogs appear in a results table. Here, you can:
- Rename or edit SMILES codes by double-clicking cells
- Click on 2D images to enlarge them
- Toggle 2D image visibility
- Generate 3D structures for selected analogs
- Remove selected analogs or clear the entire results table
From 2D to 3D Structures
To analyze analogs in greater depth, generate their 3D conformers directly via the Convert to 3D button. This sets you up for further studies, like docking simulations using tools such as the Autodock Vina Extended extension.
Why You Might Want to Try This
Whether you’re optimizing leads or exploring SAR, positional analogue scanning is a valuable shortcut to systematic molecule optimization—especially when integrated directly in your modeling environment. With interactive controls, instant 2D/3D visualization, and the ability to edit results on the fly, the SAMSON SMILES Manager helps you stay focused on insights rather than repetitive edits.
To learn more, visit the full documentation: Perform Positional Analogue Scanning using the SMILES Manager.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





