When working with complex molecular models, identifying and isolating specific structural elements—like side chains, ligands, or individual residues—can be daunting. This challenge becomes even more critical when models get large, and selections based on visual inspection simply aren’t feasible.
SAMSON offers a powerful yet flexible solution through the Node Specification Language (NSL), integrated directly into both the Find command and the Document View. In this post, we focus on using NSL as a filter tool in the Document View, helping you target exactly what you need—consistently and efficiently.
Why Use the Document View with NSL?
The Document View in SAMSON is like the file explorer of your molecular model. But instead of being limited to expanding folders and checking boxes, you can filter node types by their properties using NSL. This is especially useful when you need to:
- Quickly find all side chains, backbones, or hydrogen atoms
- Filter out specific node types (e.g.,
node.type visualModel) - Focus on a range of residues based on coordinates or IDs
Using NSL in Document View
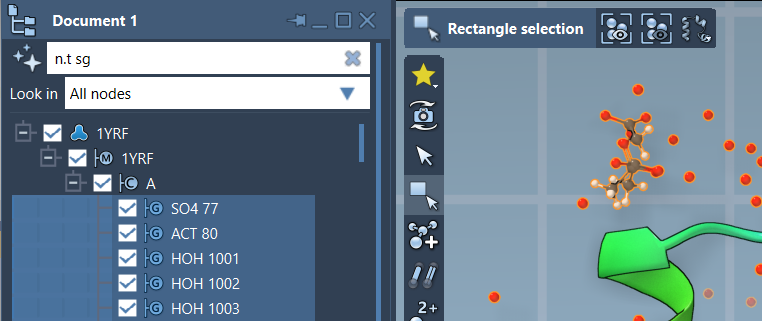
You can enter NSL expressions directly in the Document View filter. For example, the query:
|
1 |
n.t sg |
short for node.type structuralGroup, filters the view to show only structural groups. Press Enter to apply the filter and select the visible nodes.
The NSL engine is sensitive to node hierarchies, and with proper queries, you can filter by chemical or structural attributes, like:
node.category ligand, receptor: shows ligands and receptorsresidue.secondaryStructure helix: isolates alpha helicesn.t sc h S: filters side chains that have sulfur atoms
Let AI Assist You
If you’re not sure about the expression syntax, SAMSON includes an AI Assistant right next to the filter box. It understands your model and can generate an NSL expression tailored to your document’s structure. Ideal for beginners and time-saving for experts.
Tips to Improve Your Filtering Workflow
- Use quotes to select nodes by name:
"ALA 22" - Wildcard helps broaden selections:
"ALA*"includes all nodes starting with “ALA” - Use logical operators:
n.t r and not r.t ALAfilters out alanine residues
Wrapping Up
The integration of NSL in the Document View gives you granular control over your molecular models. Small improvements in your selection process can dramatically multiply your productivity, whether you’re preparing simulations or generating visuals.
To learn more and explore even more examples and use cases, visit the official NSL documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. Get SAMSON at https://www.samson-connect.net.





