Setting up lipid membranes around proteins is a recurring task in molecular modeling and simulations, particularly for those working on membrane proteins or studying membrane-protein interactions. However, assembling lipid layers manually can be tedious and time-consuming. Fortunately, SAMSON’s Molecular Box Builder offers a reproducible and visual way to populate your simulation box with lipids, right around your protein of interest.
This blog post walks through how to use Molecular Box Builder to create a lipid layer—or even a bilayer—around a membrane protein in SAMSON, using a real example. The process is entirely visual, and gives you flexibility regarding orientation, sizing, and lipid selection. If you have ever hesitated at the command-line pile-up of membrane setup, this may be a useful alternative.
Reorienting the Protein
Before adding lipids, it’s important to orient the protein in the intended direction—typically aligning its transmembrane axis with the Z-axis. Here is how:
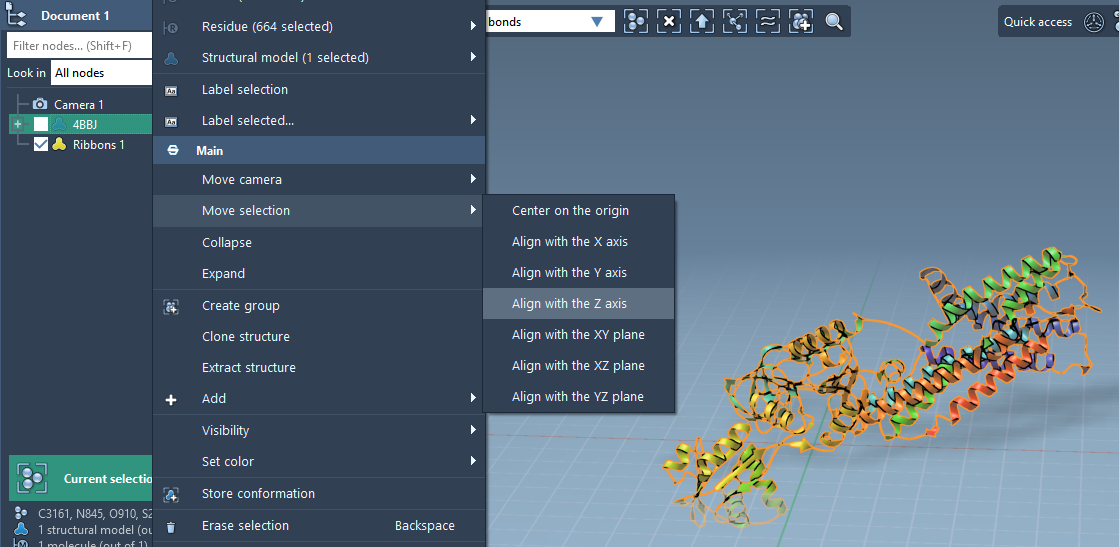
- Right-click the protein in the Document view.
- Choose Move selection > Align with Z axis from the context menu.
- Then choose Move selection > Center on the origin.
Select and Align the Lipid
Next, import your lipid molecule. This could be POPC or any other type appropriate for your study. Once loaded:
- Select the lipid and open the Molecular Box Builder app.
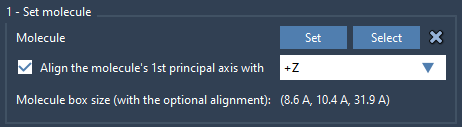
- Click Set to define it as the molecule to be inserted.
- Choose to align its principal axis with the
+Zdirection (matching the orientation of the protein).
Define Your Insertion Box
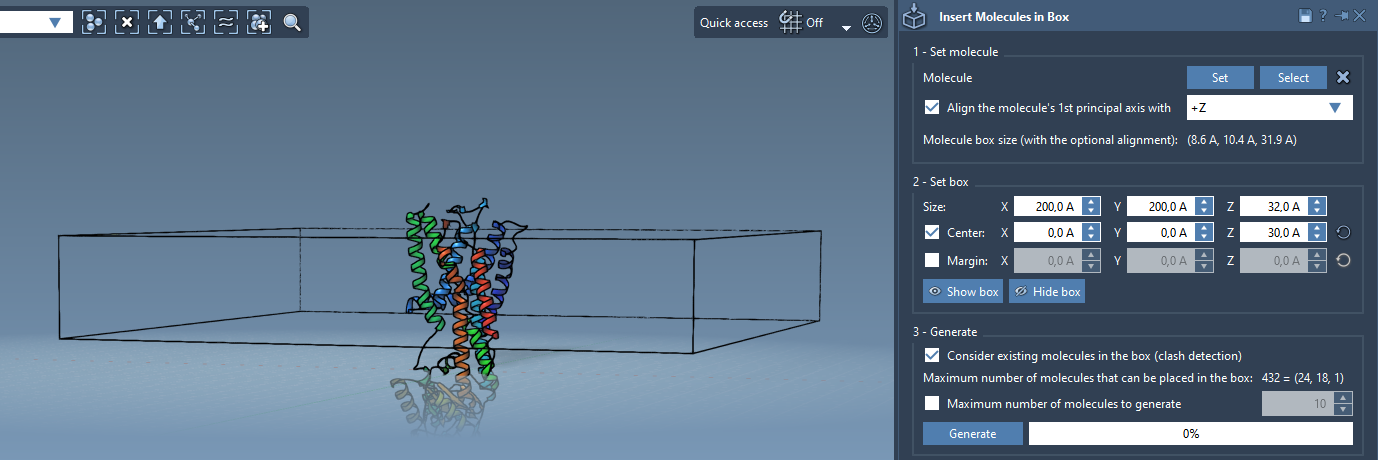
Now, we set up a box around the protein, into which lipids will be inserted:
- Center the box on the protein.
- Adjust the box dimensions so it fits a single lipid layer (typically narrow on Z, wider on X and Y).
- Optionally, set the margin between lipids to fine-tune spacing.
Generate the Layer
Before clicking generate, be sure to check Consider existing molecules in the box. This ensures lipids are only placed where there is space—i.e., around the protein but not inside it.
- Click Generate to fill the box with non-overlapping lipids around the protein.

Optional: Add a Second Lipid Layer
To build a bilayer, simply:
- Add the first layer using
+Zalignment. - Shift the insertion box center downwards in Z.
- Add the second layer with
-Zalignment and click Generate again.
This flexibility allows you to construct symmetric or asymmetric bilayers directly around your protein model.
Lipid layers can then be solvated and prepared for simulation using tools like GROMACS Wizard, which integrates seamlessly within SAMSON. This approach offers a straightforward, visual alternative to scripting-based methods.
To learn more about how to use Molecular Box Builder, visit the full documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





