Designing new molecules often involves tweaking known structures – replacing parts of a molecule to generate analogues for screening. This process can be both time-consuming and error-prone when done manually, especially at scale. If you’re working in drug design, materials science, or chemical biology, you’ve likely faced the challenge of generating diverse libraries with small structural modifications, while preserving core functionalities.
The SMILES Manager extension in SAMSON offers an efficient way to do this via the Replace fragments tab. It provides chemists and molecular designers with a fast, visual, and customizable approach to perform fragment-based molecule generation – all without scripting or leaving the SAMSON interface.
What the Replace Fragments Tool Does
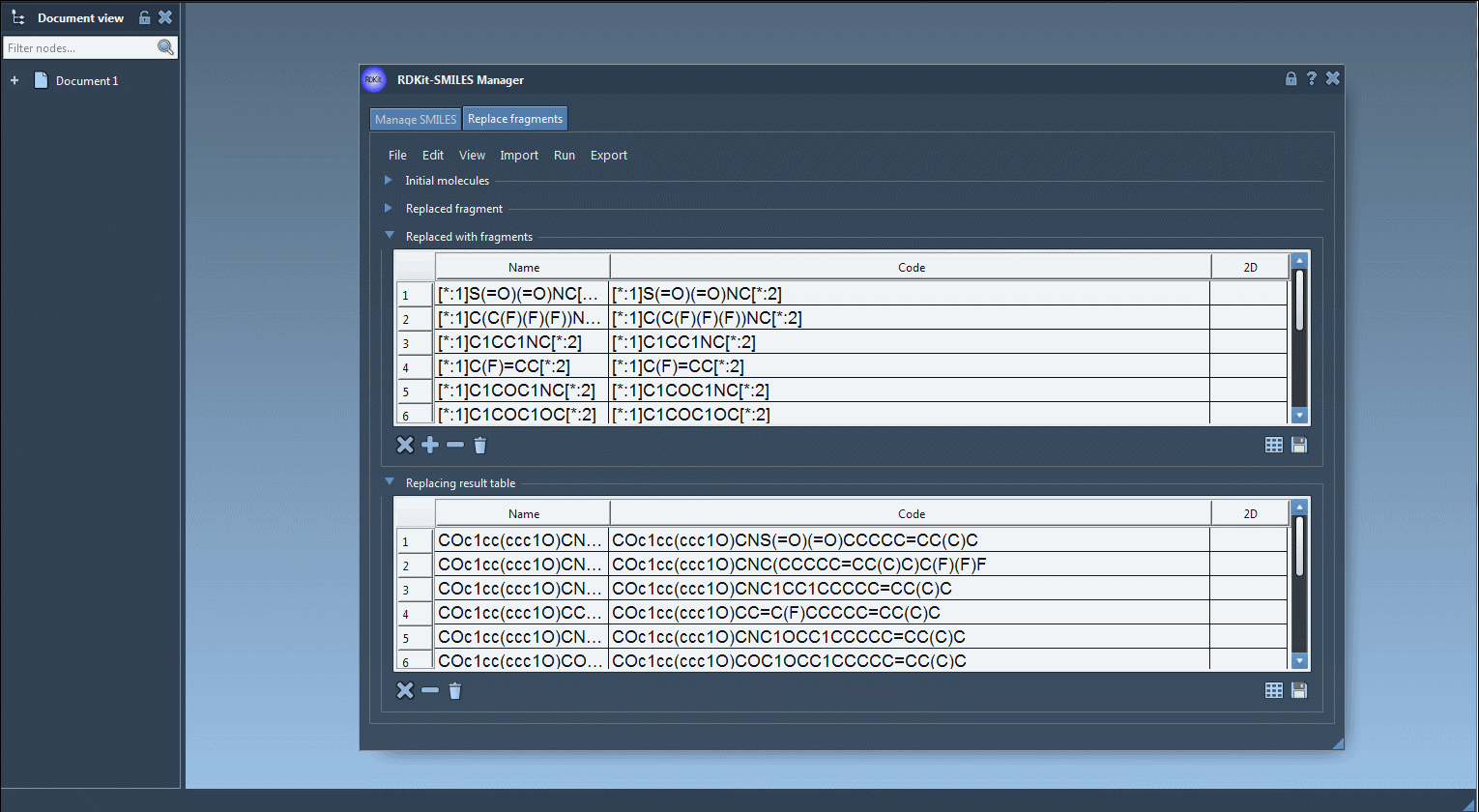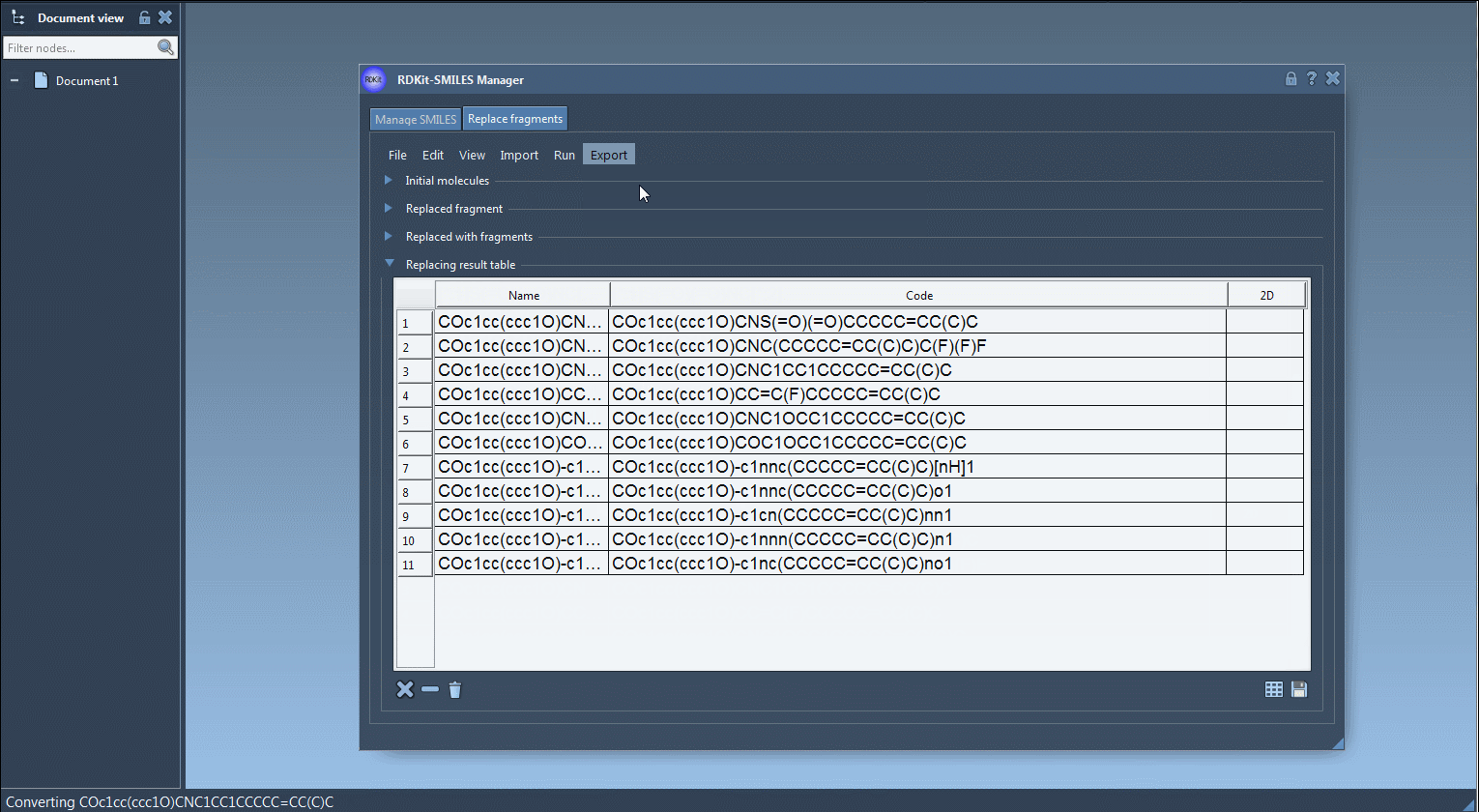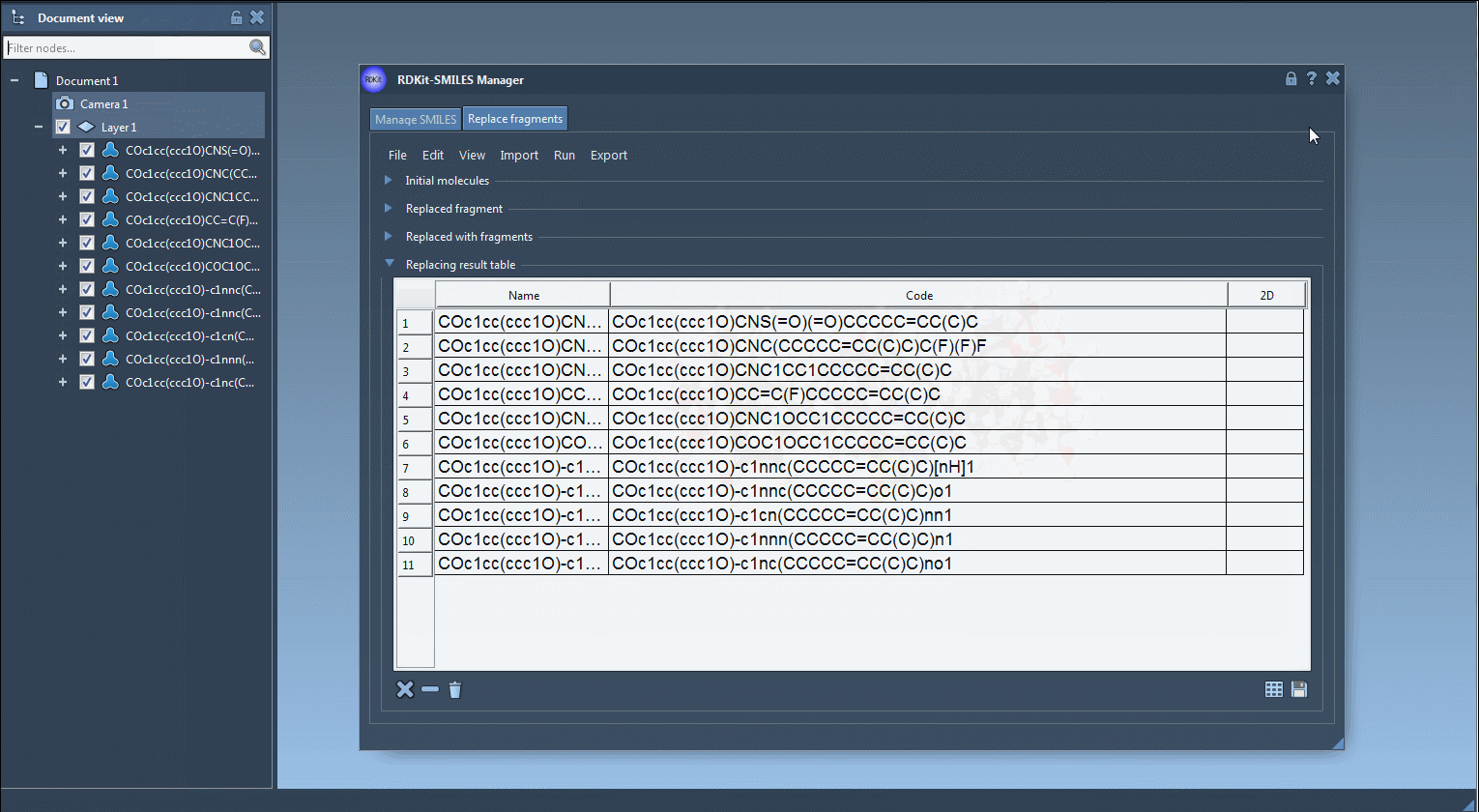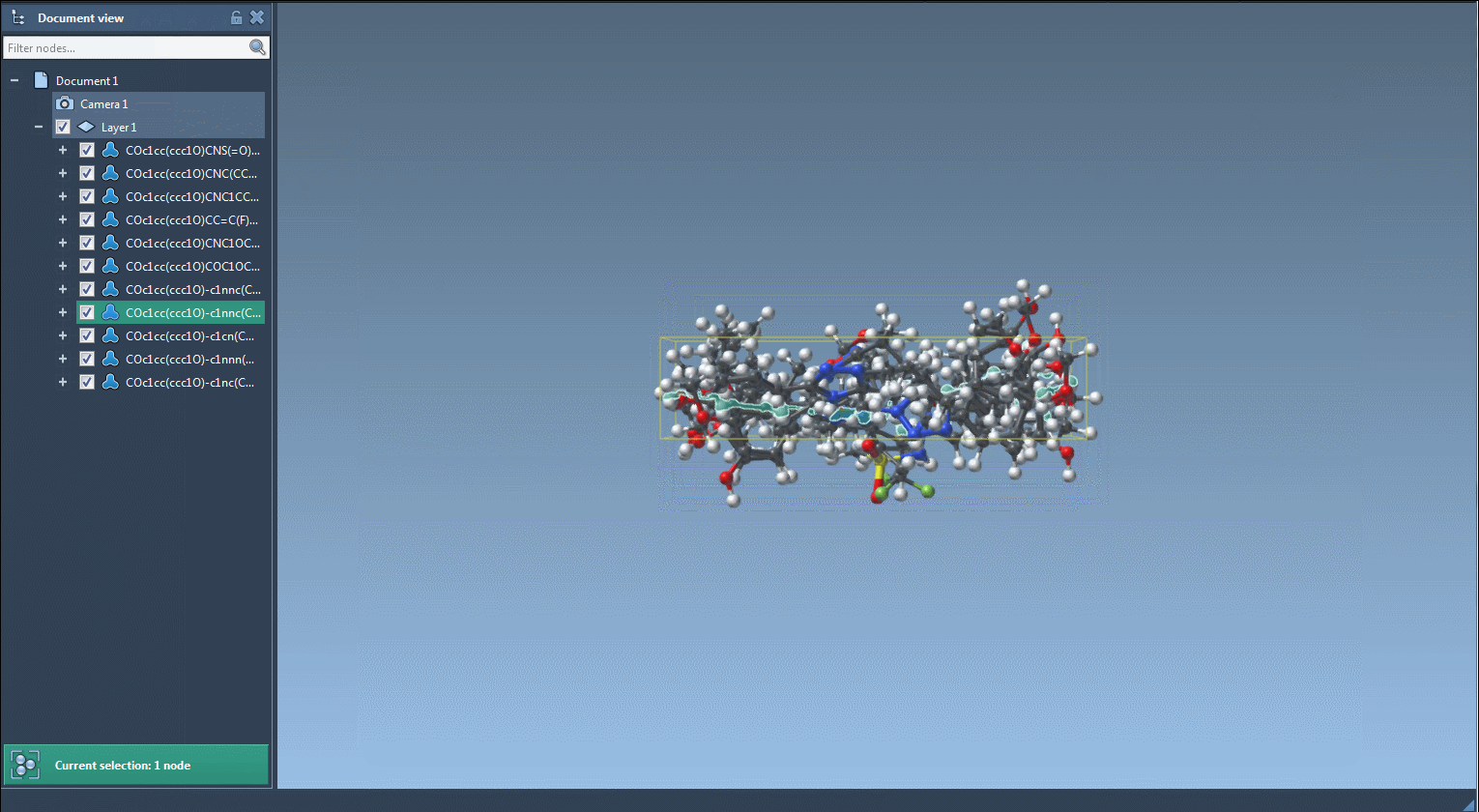
The Replace fragments tab allows users to define one or more starting molecules, specify a target fragment pattern, and then substitute it with different fragments. This is particularly helpful for generating isosteric libraries or exploring SAR (structure–activity relationships).
Fragments are defined using SMARTS patterns like [*:1]C(=O)NC[*:2], where [*:1] and [*:2] denote variable attachment points. You can import these patterns from a text file, enter them manually, or use your current selection inside a SAMSON document.
Example: Replacing an Amide Linker
Let’s walk through an example to generate analogues for the molecule:
|
1 |
COc1cc(CNC(=O)CCCC/C=C/C(C)C)ccc1O |
Suppose we want to replace the amide linker C(=O)NC with other functional groups. Here’s how you’d do it:
- Load the original molecule into the table via file import or by pasting its SMILES string.
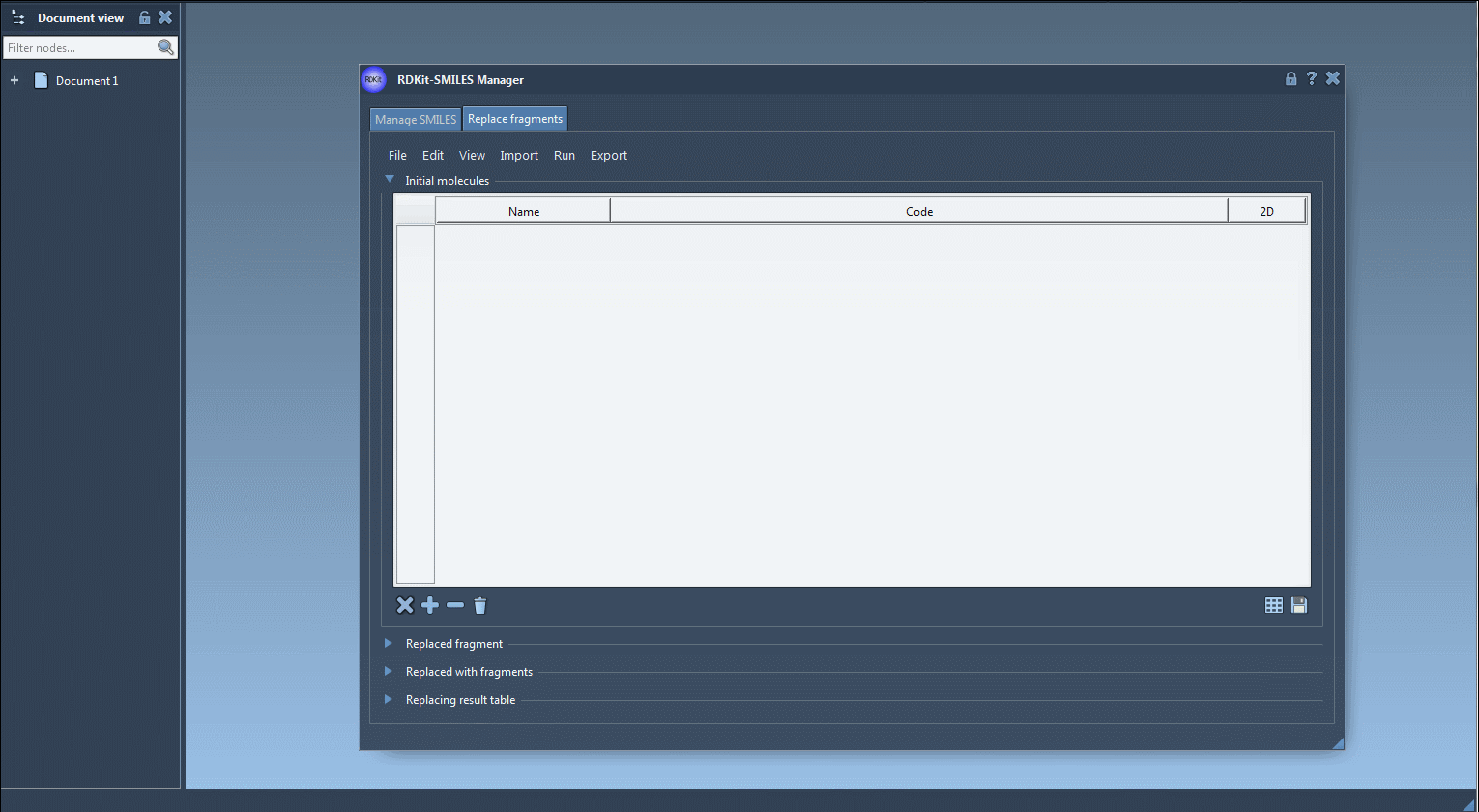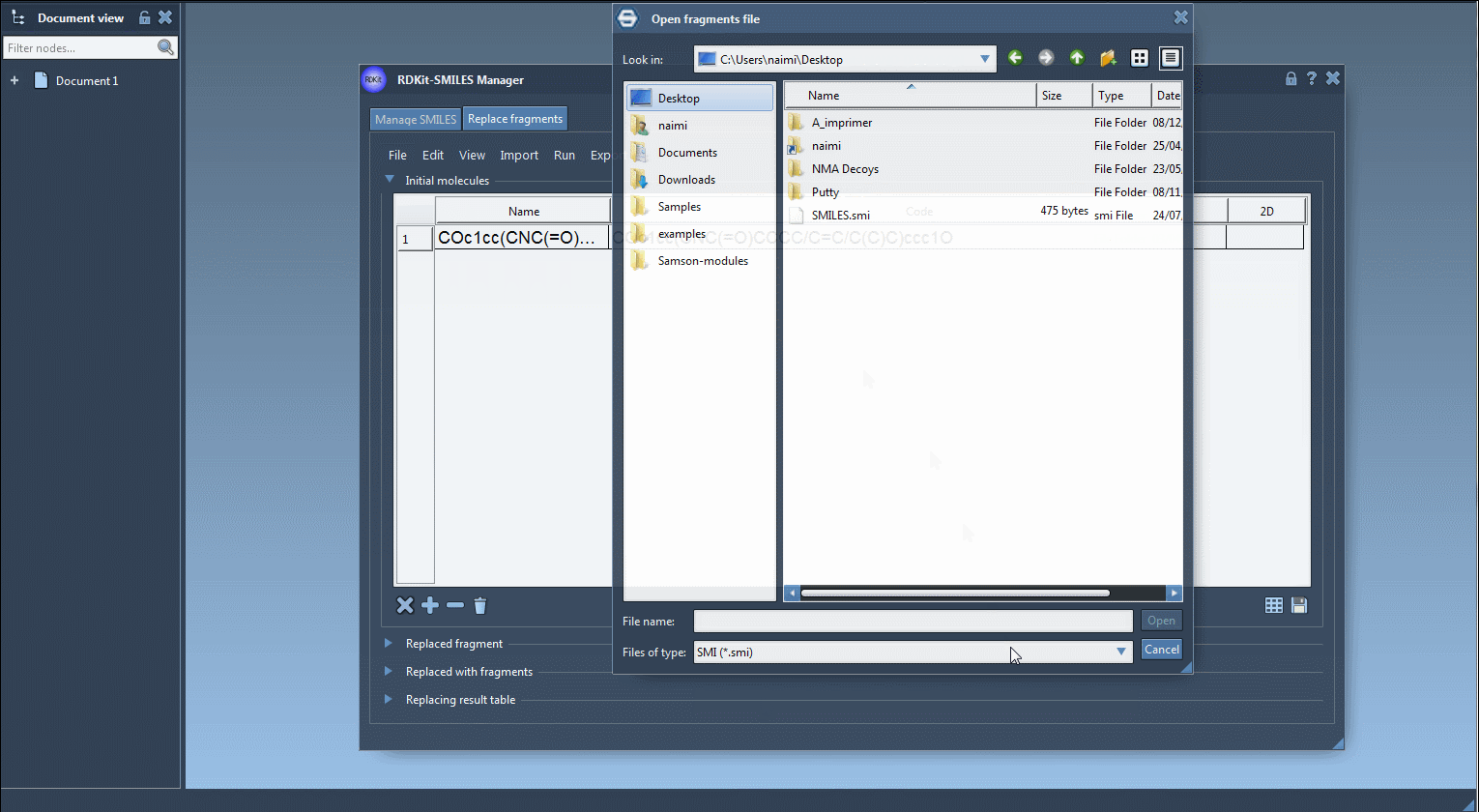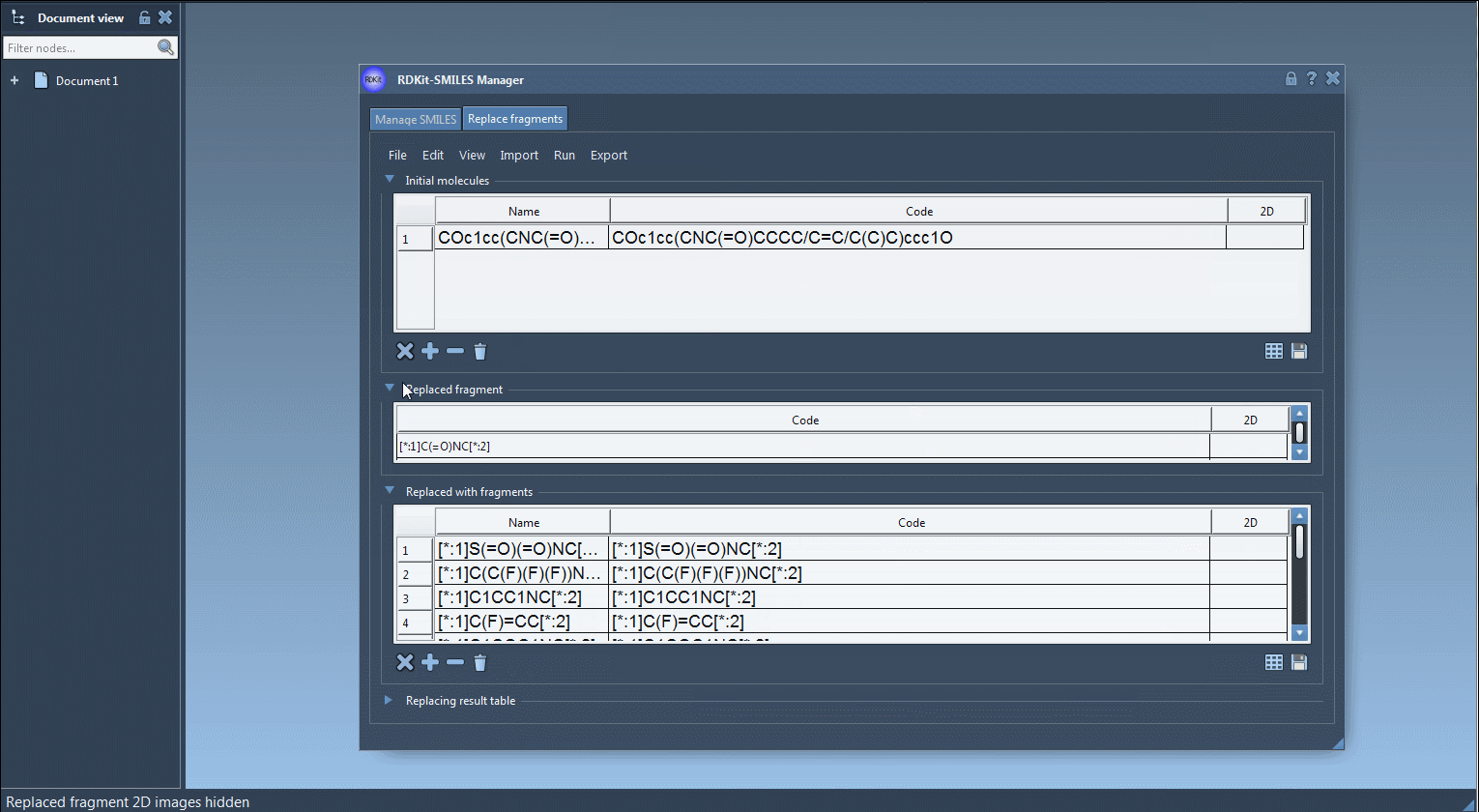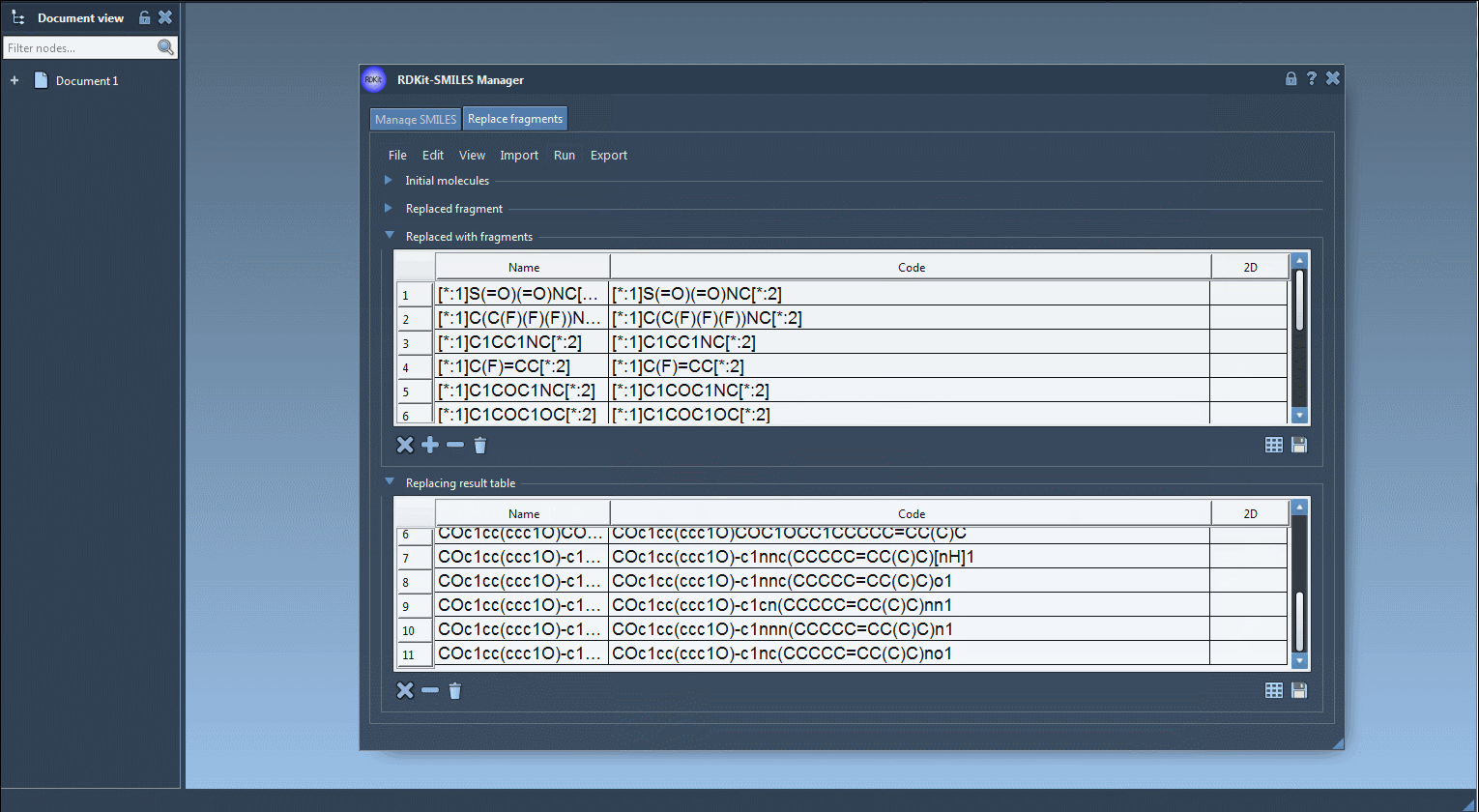
- Specify the fragment to replace:
[$([*:1]C(=O)NC[*:2])] - Paste the replacement fragments into the table (e.g.,
[$([*:1]S(=O)(=O)NC[*:2])],[$([*:1]C(C(F)(F)F)NC[*:2])], etc.) - Click on Run > Replace fragments
- View the results in the output table — each row is a new molecule.
Each new structure can then be converted automatically into a 3D conformation, visualized in SAMSON or exported.
Why It Matters
This feature simplifies analog generation significantly, allowing you to focus on evaluating properties rather than drawing molecules. It eliminates the need to manually script SMILES manipulation or juggle spreadsheets and cheminformatics libraries. Moreover, it enables iterative design in an interactive environment where you can visualize changes instantly.
What You Can Do with the Results
- Visualize 2D and 3D structures instantly.
- Export the molecules to your active document or save them as individual files or grid images.
- Perform further analysis using other SAMSON extensions for docking, property prediction, etc.
A Few Tips
- Make sure fragments you use for substitution have correct attachment points (
[*:1]and[*:2]). - You can use other powerful SAMSON features with the generated molecules, such as energy minimization or advanced visualization tools.
If you often build analog libraries, this can save hours of work and streamline your workflow. Explore more in the full SMILES Manager documentation:
Learn more in the documentation
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





