When studying molecular interactions, especially those involving binding or unbinding events, researchers often need access to atomic coordinates along well-defined paths. These trajectories are useful for reaction coordinate analysis, free energy calculations, or simply visualizing how a ligand travels through a protein structure.
However, many molecular modelers struggle to extract just the relevant movement data in a structured format that can be used downstream—for instance, in analysis tools or simulation packages. Manually tracking atom positions across frames is tedious and error-prone. The Export Along Paths extension in SAMSON offers a flexible and user-friendly way to export the coordinates of atoms—either all atoms or a selected subset—along a previously defined path. In this post, we explain how to export the trajectory of a specific group of atoms (such as a ligand) along a path.
Exporting Only the Atoms You Care About
If you’ve already computed ligand trajectories using, for example, the Ligand Path Finder extension, the Export Along Paths app allows you to export just the ligand atoms—letting you focus only on what you need. Here’s a step-by-step guide to doing this.
Step 1: Load the Sample System
To try it out easily, download this sample document from SAMSON Connect. It includes the membrane protein Lactose permease (PDB: 1PV7) with its ligand Thiodigalactosid (TDG) and precomputed unbinding paths.

Step 2: Launch the Export Tool
Open the Export Along Paths app via Home > Apps > All > Export Along Paths.
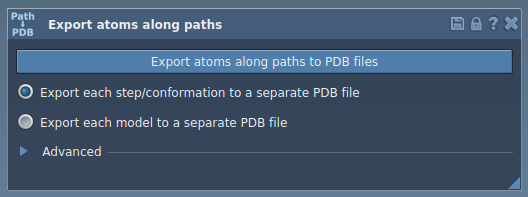
Step 3: Select and Export a Subset of Atoms
- Expand the Advanced panel in the Export app interface.
- Use the Document View to select only the ligand atoms (e.g., TDG).

- Click Add to register the selection as a model for export.
This will add the selected group of atoms as an exportable model. You can rename this group and add more selections if needed.

When you’re ready:
- Select a path in the Document View (this is the trajectory you want to export).
- Choose an export format: either a single multi-frame PDB or multiple PDBs (one per frame).
- Click Export atoms along paths to PDB files and choose the output folder and file prefix.
When Is This Useful?
Exporting a subset of atoms along a path is particularly helpful in several cases:
- Generating reaction coordinate files for free energy calculations.
- Extracting ligand trajectories for enhanced sampling simulations.
- Visualizing how specific molecules interact with a binding pocket over time.
Unlike exporting everything, focusing just on what matters can save computation time downstream and keeps your data organized.
To learn more and see the full documentation, visit the Export Along Paths documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON here.





