Designing molecular analogues is a common approach in medicinal chemistry to explore structure-activity relationships and optimize compounds. However, scanning and editing molecular substructures manually can be both slow and error-prone. If you’re looking for a way to streamline analogue generation using a reproducible and structured approach, positional analogue scanning using the SMILES Manager in SAMSON may help you get there faster and more systematically.
In this post, we’ll walk you through how to perform targeted substitutions on specific molecular patterns directly within the SMILES Manager, using simple SMARTS queries and intuitive menus. This process can help you quickly explore the chemical space around your molecule of interest.
Identifying a Replaceable Pattern
The first step in this process is to select the atoms or patterns you’d like to scan for potential substitutions. In SAMSON, this is done by entering a SMARTS pattern — a flexible way to define chemical substructures. For example, to target an aromatic carbon bearing a hydrogen atom, you can use the SMARTS pattern [cH].
Using this query, SMILES Manager will highlight all matching atoms in your molecule. This visual check ensures clarity on which parts of your structure will be considered for modification.

Making Targeted Changes
Once a substructure pattern is identified, you can decide how to modify it. SMILES Manager offers two main strategies:
- Replacement: Replace a matched atom with another atom (e.g., swap a carbon for a nitrogen).
- Attachment: Add a group or atom to the matched site (e.g., attach a fluorine or methyl group).
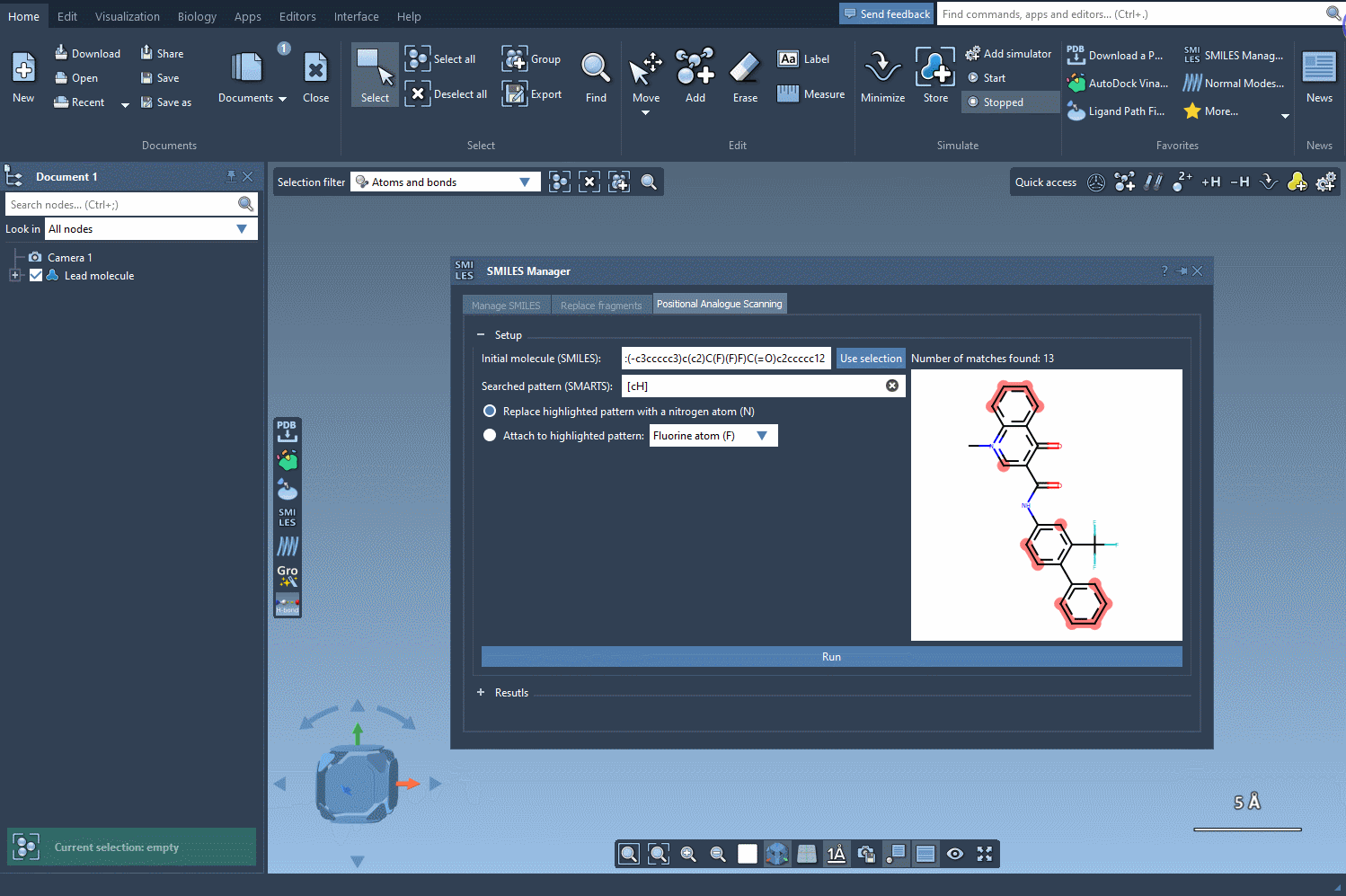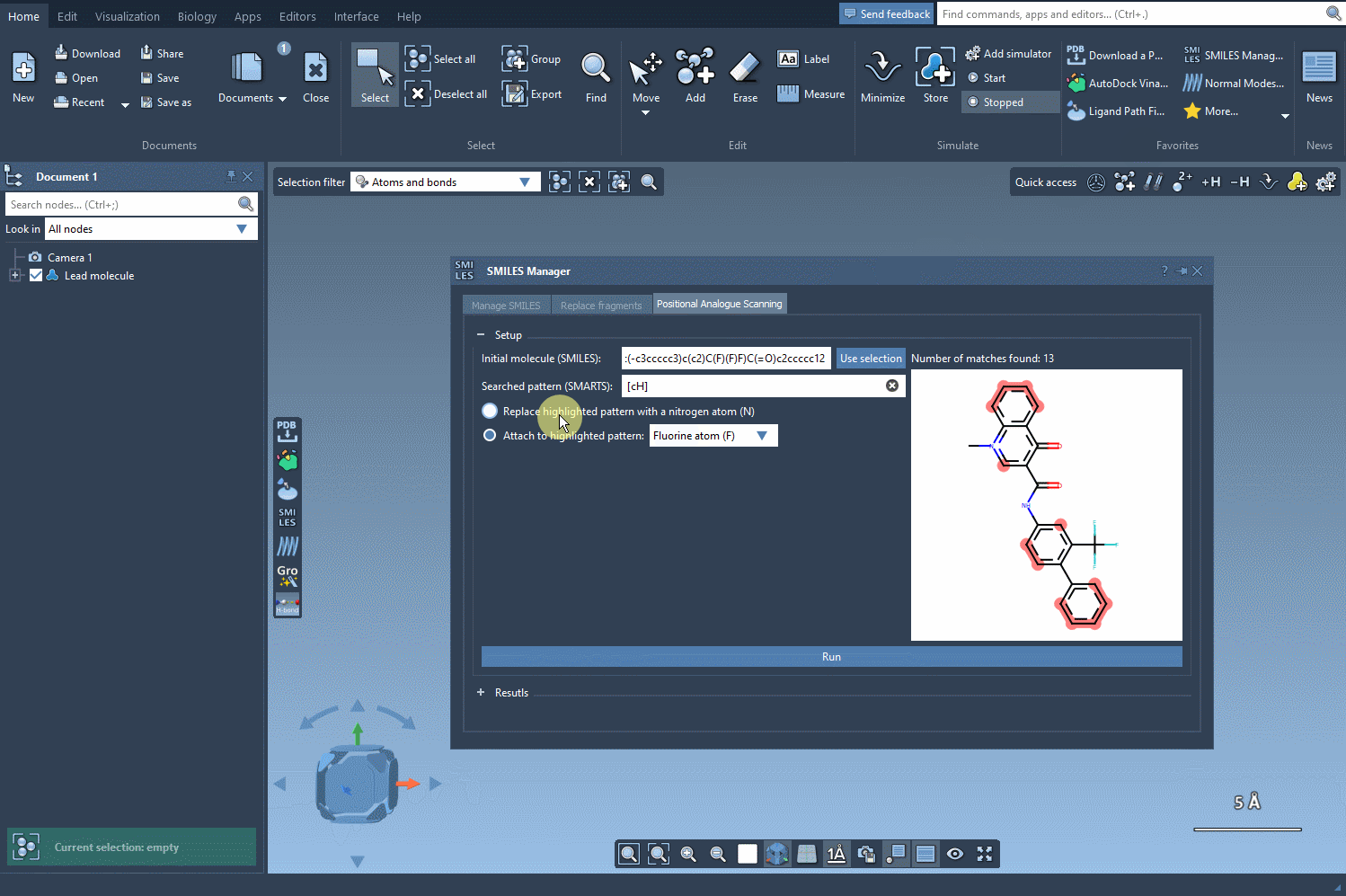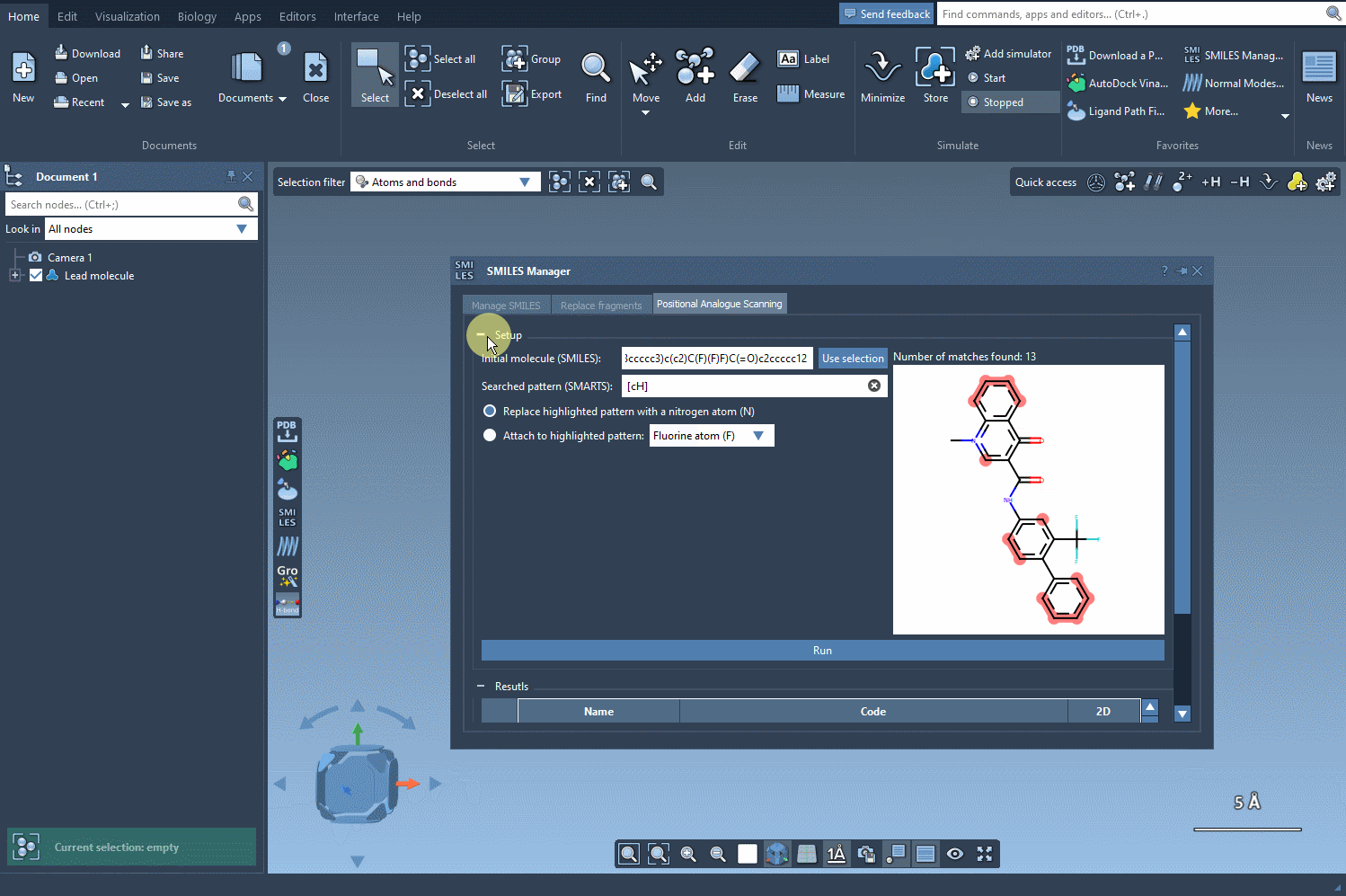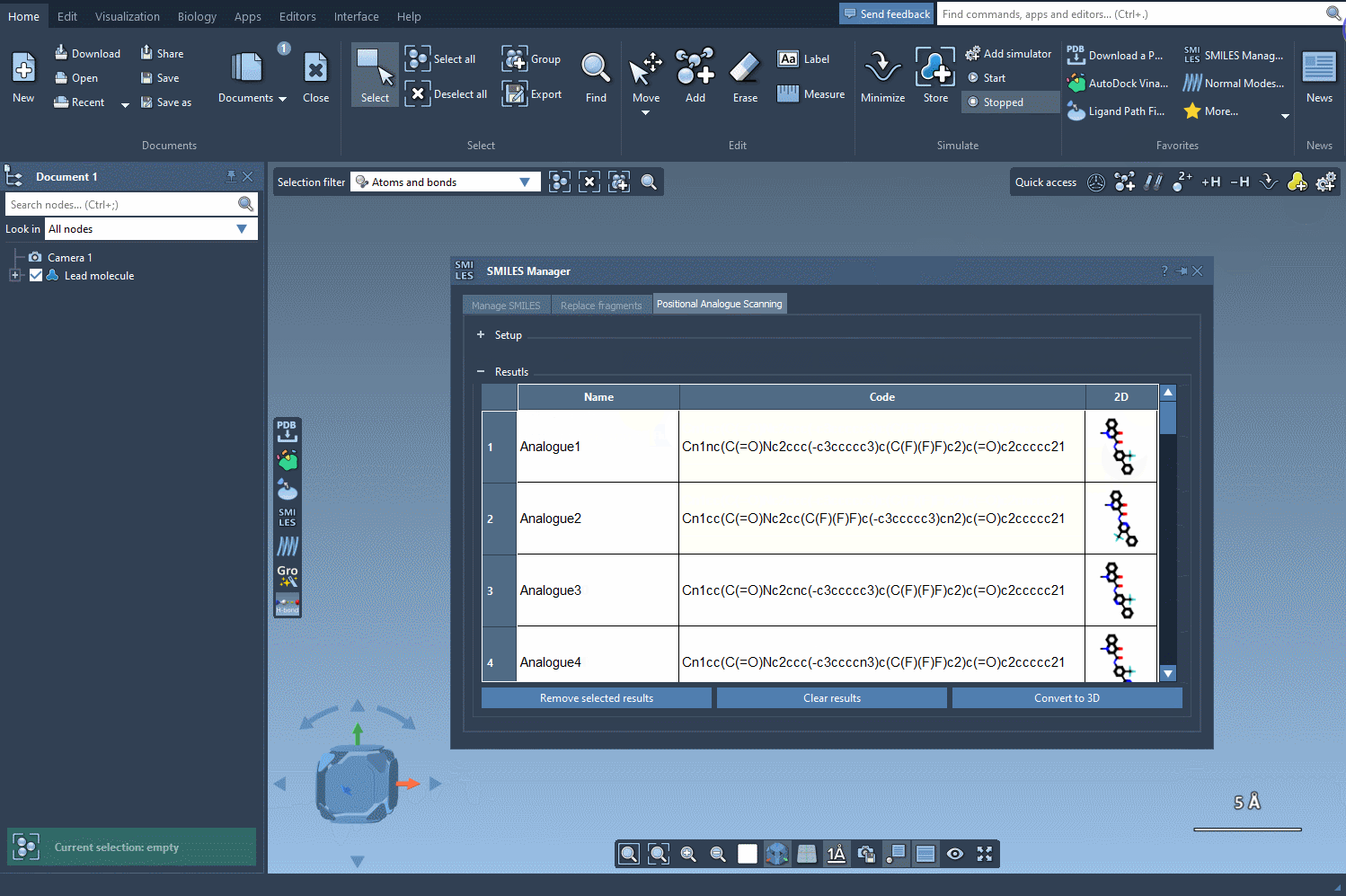
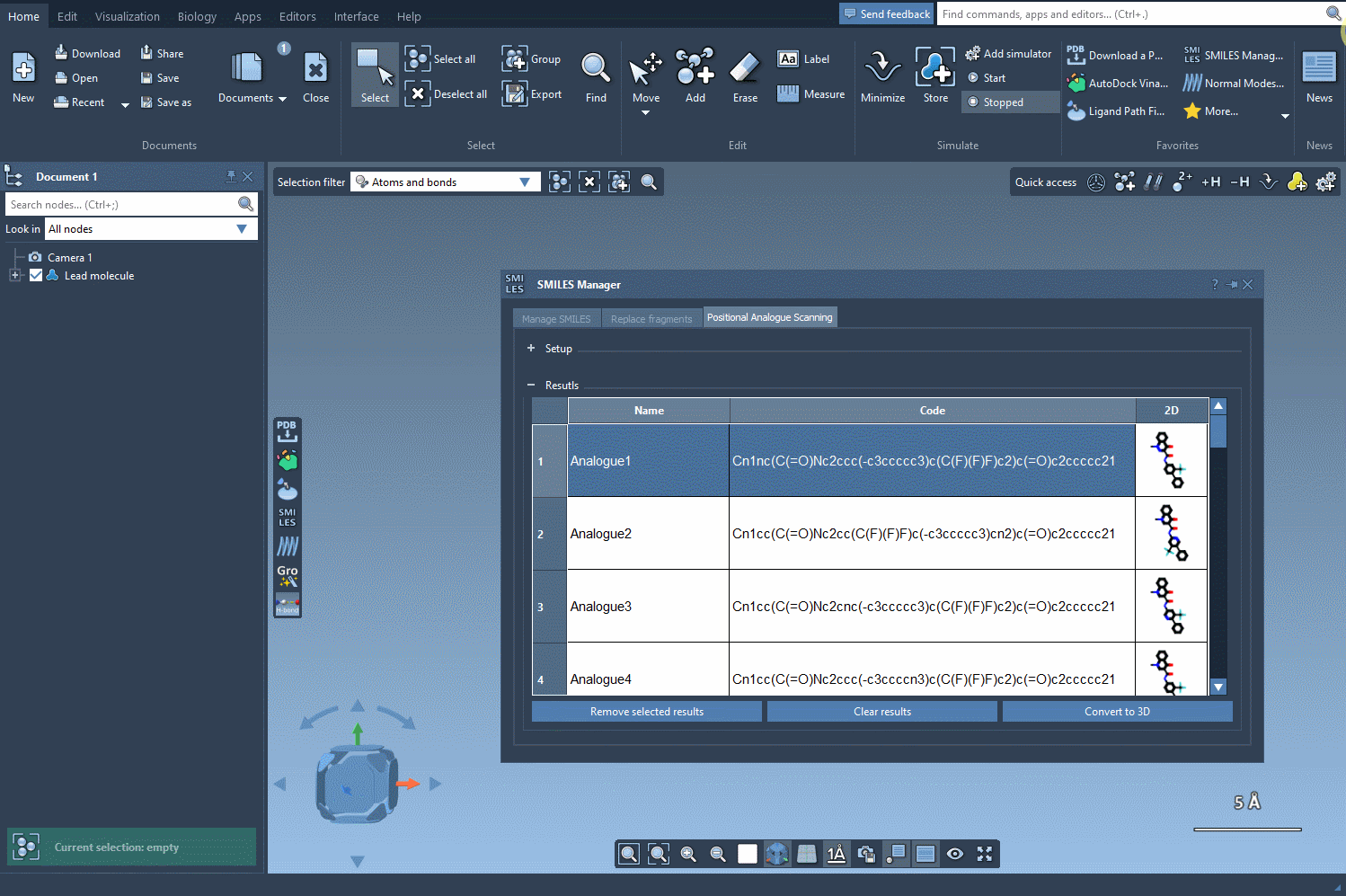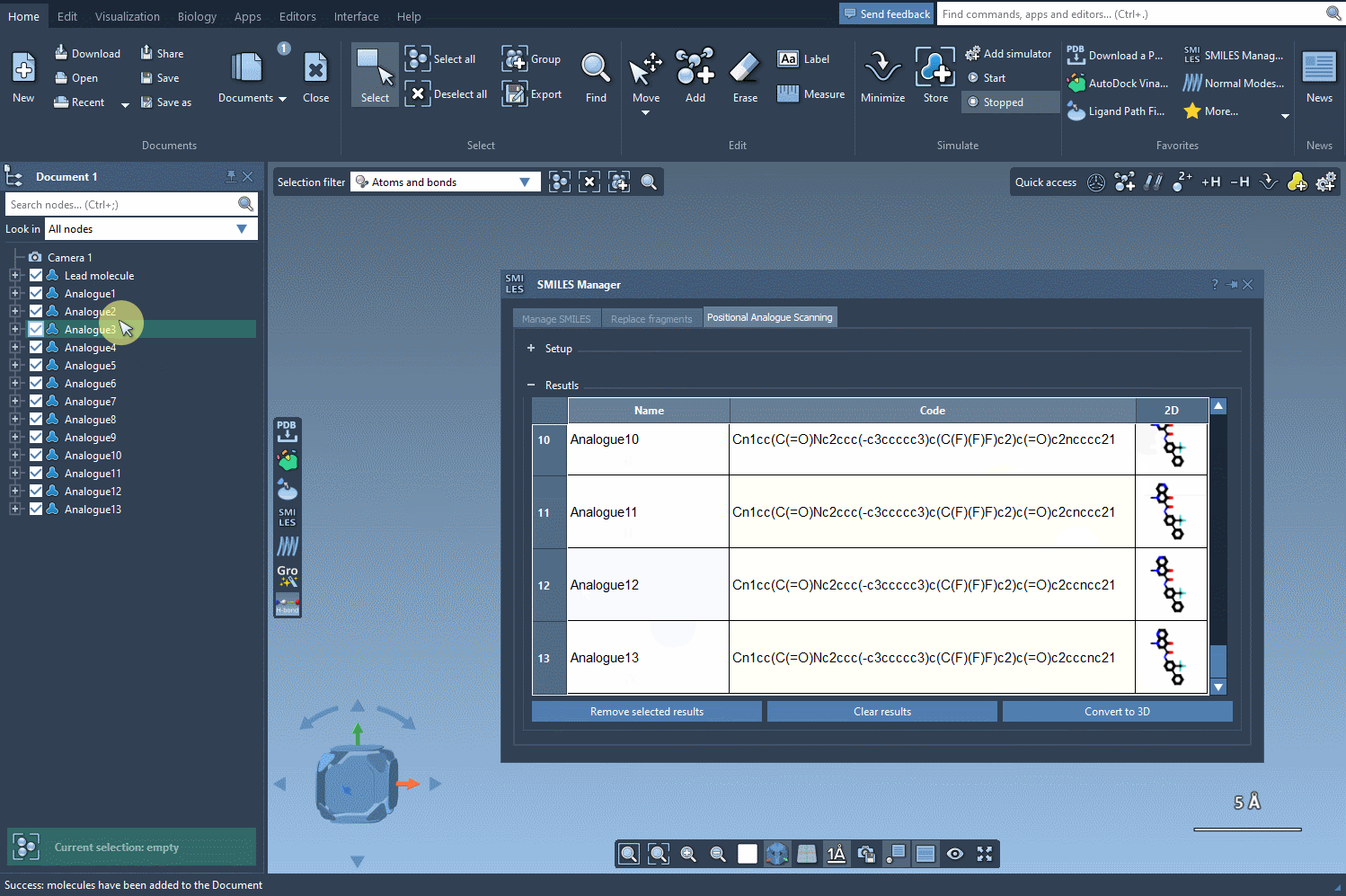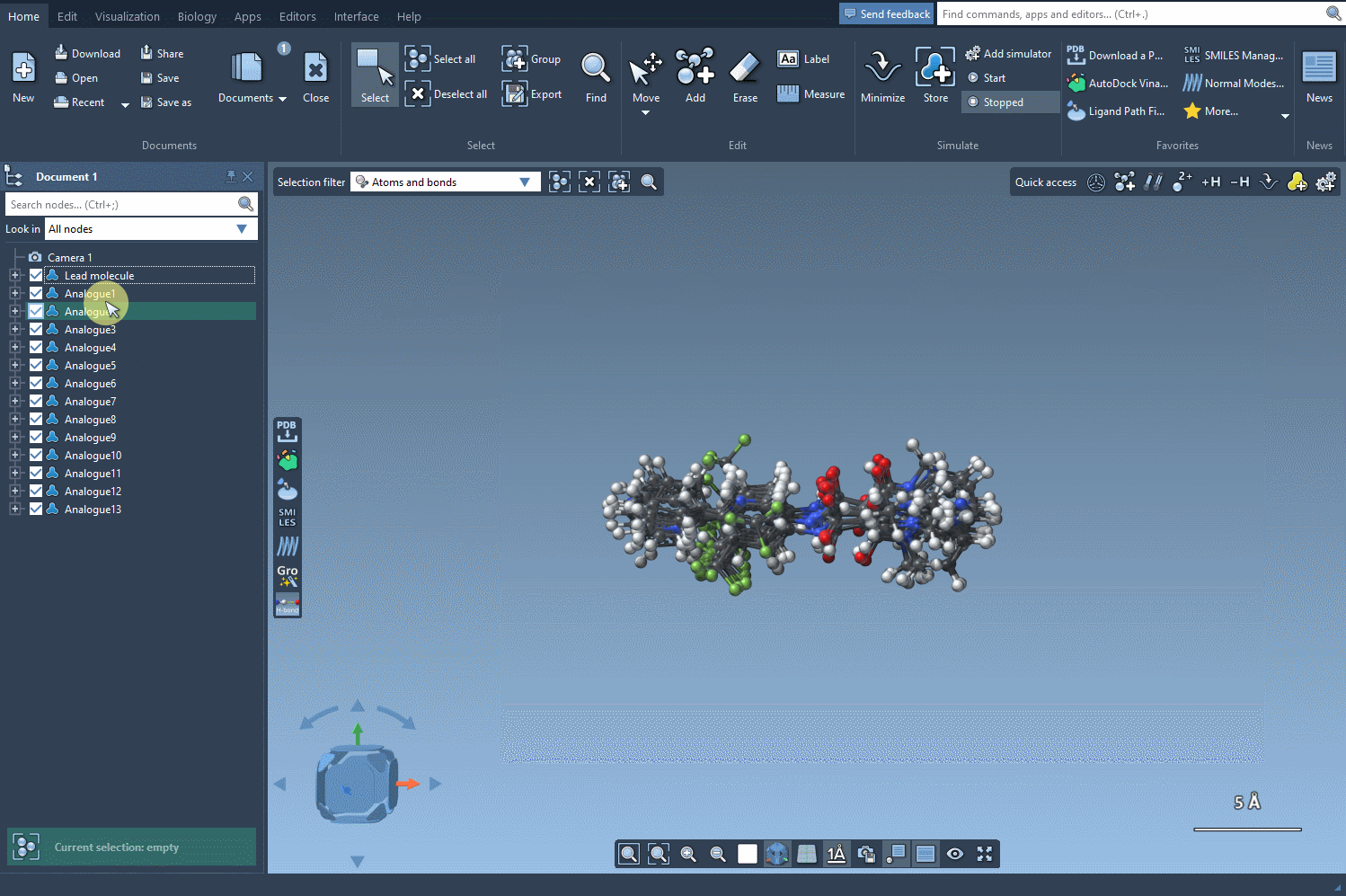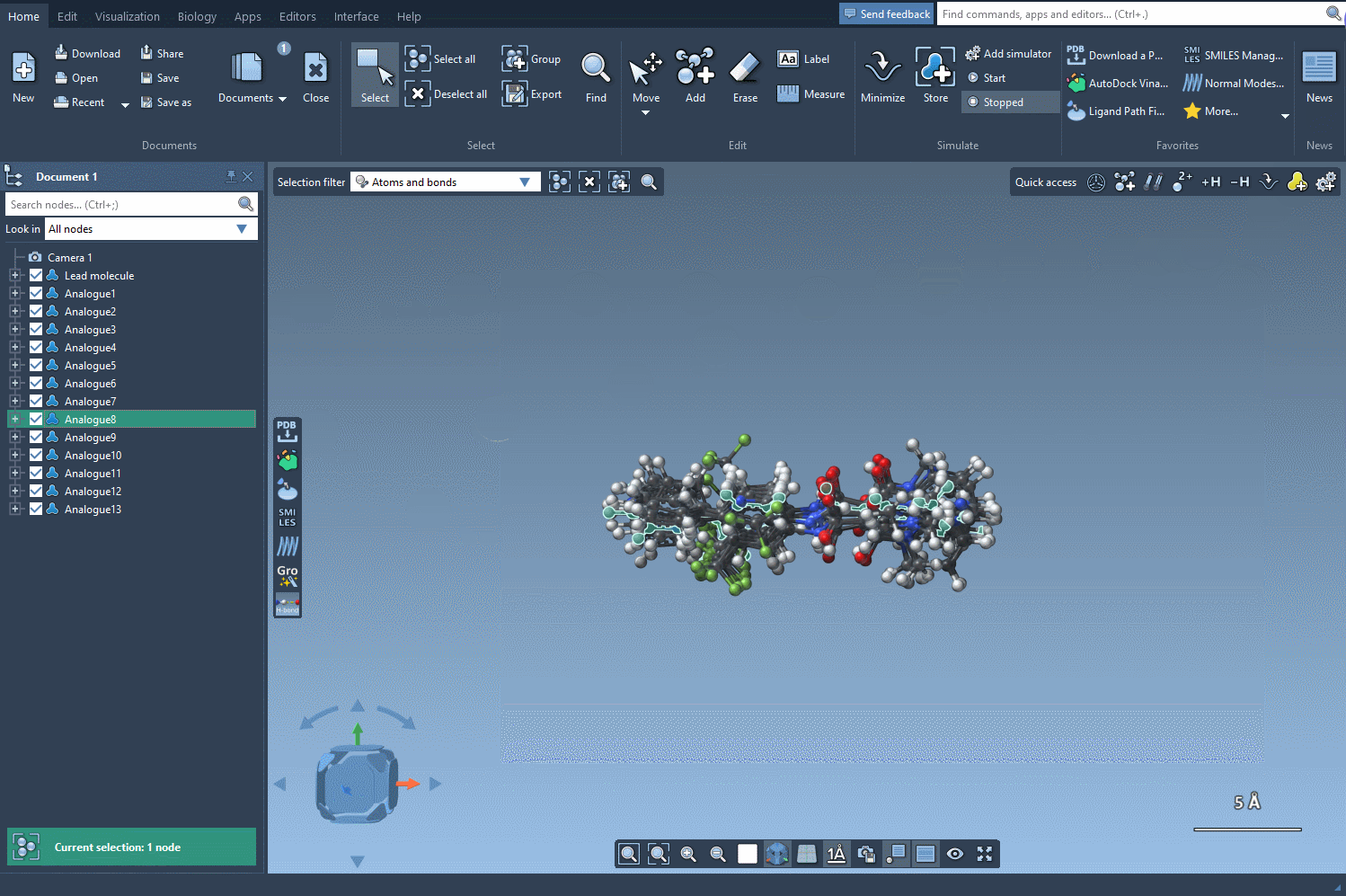
You can choose from a set of common modifications—especially useful in early-stage medicinal chemistry iterations. Once you’re ready, hit Run, and the analogues will be rapidly generated and visualized with 2D depictions in the results table.
Why Bother with SMARTS?
If you’re wondering why this isn’t just handled manually, consider how much time is saved when exploring tens or hundreds of variations. SMARTS queries ensure consistent, automated identification of substructures, and reduce human error in multi-step analogue synthesis planning. This clean, reproducible process fits naturally in a workflow that integrates computation and medicinal chemistry design.
From Sketch to 3D
The analogues you generate aren’t just structural placeholders. You can directly convert the 2D SMILES into 3D structures using the Convert to 3D button within the results table.
From here, you can run downstream analyses such as docking using Autodock Vina Extended or visualize interactions with target proteins — all within the same unified workspace in SAMSON.
Conclusion
Whether you’re early in a lead optimization process or trying to generate hypotheses based on structure-activity gaps, positional analogue scanning offers a fast and rigorous way to explore variations. It takes just a few clicks, thanks to integrated SMARTS support and automatic 3D generation — saving time while ensuring chemical plausibility.
To learn more, visit the full tutorial page here: https://documentation.samson-connect.net/tutorials/smiles-manager/perform-positional-analogue-scanning-using-the-smiles-manager-element/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





