When studying molecular mechanisms—such as ligand unbinding or chemical reactions—it’s often not enough to know just the starting and ending molecular structures. Molecular modelers need to understand the transition path a molecule follows between states, particularly through areas around the saddle point—the highest energy point along the minimum energy path. This type of data can guide drug design, enzyme engineering, or even materials development. But calculating these paths with sufficient accuracy and speed can be tedious and computationally expensive.
This is where the Parallel Nudged Elastic Band (P-NEB) app in SAMSON comes in. P-NEB improves and automates the optimization of transition paths using a parallelized version of the widely-used Nudged Elastic Band (NEB) method. This blog post focuses on how to improve transition paths between conformations using P-NEB, based on a practical example that’s both accessible and informative for new users.
Why Use P-NEB on Transition Paths?
In simple terms: accuracy and control. P-NEB helps refine pre-computed pathways—such as those generated by linear interpolation or higher-level methods like SAMSON’s Ligand Path Finder—to better represent real energy landscapes. The algorithm minimizes each intermediate image along the path while preserving equal spacing, which results in more realistic and physically meaningful transitions.
Let’s Try It: Improving a Ligand Unbinding Path
SAMSON provides pre-made models to get started. One such example includes the unbinding path of a zinc ligand:
https://www.samson-connect.net/documents/39260103-9a48-49db-9488-5573d5f1e7b0
This serves as an ideal lightweight test system. After downloading the document through Home > Download in SAMSON, you can launch the P-NEB app via:
Home > Apps > All > P-NEB
![]()
P-NEB Settings: What to Use and Why
The app interface asks for several parameters:
- Spring Constant: Set this to
1.00. - Number of Loops: A good starting point is
100. - Interaction Model: Use the
Universal Force Field (UFF). - Optimizer: Choose
FIRE(Fast Inertial Relaxation Engine). - Parallel Execution: Check this for faster computation.
- Climbing Image: You can leave this unchecked initially.
- Suffix Name: Use something descriptive, like
NEB.
Applying P-NEB to a Path
1. In the Document view, select a path node.
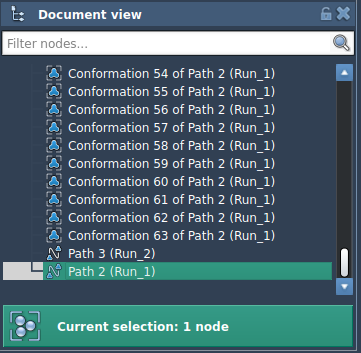
2. Click Run in the P-NEB app.
3. The app prompts you whether to use existing bonds—select ‘Yes’.
4. Computation begins. You can track it via the status bar:

Once complete, a new path will appear in your document with optimized transition frames.
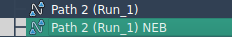
What Next?
You can now explore the new path dynamically—animate it, inspect it frame-by-frame, and use it for further analysis or visualization. This workflow is particularly useful when optimizing unbinding events in drug targets, or when trying to better understand rare transitions in molecular systems.
To go further, consider comparing this optimized path to one created from a manually generated set of conformations, or toggle on the climbing image option to refine saddle point detection.
🧪 Useful Tip: If you have only a set of conformations, you can convert them into a path using:Conformation > Create path from conformations
To learn more or follow an in-depth tutorial, visit the full documentation page:
Optimize transition paths with the Parallel Nudged Elastic Band method
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





