When working with protein structures from the Protein Data Bank (PDB), you’ll often find yourself looking at what’s called the asymmetric unit. This is just one portion of the complete biological assembly — the full, functional form of the protein, often symmetrical and made up of multiple chains.
For molecular modelers and structural biologists, it’s critical to reconstruct the full assembly when analyzing interactions, building functional oligomers, or setting up simulations. But that can be a challenge when the original PDB file only provides part of the picture.
The Symmetry Mate Editor in the SAMSON molecular design platform helps solve this problem by using transformation data directly from PDB files to regenerate symmetry mates. This means you can virtually “rebuild” the full crystal structure or biological assembly without manual alignment or exporting chains into external tools — all directly inside your modeling environment.
Visualize Symmetries — Interactive and Real-Time
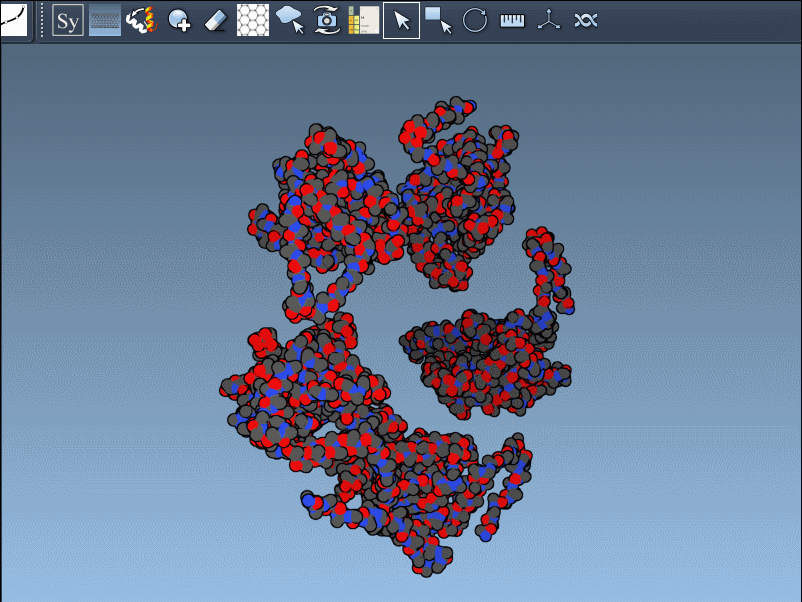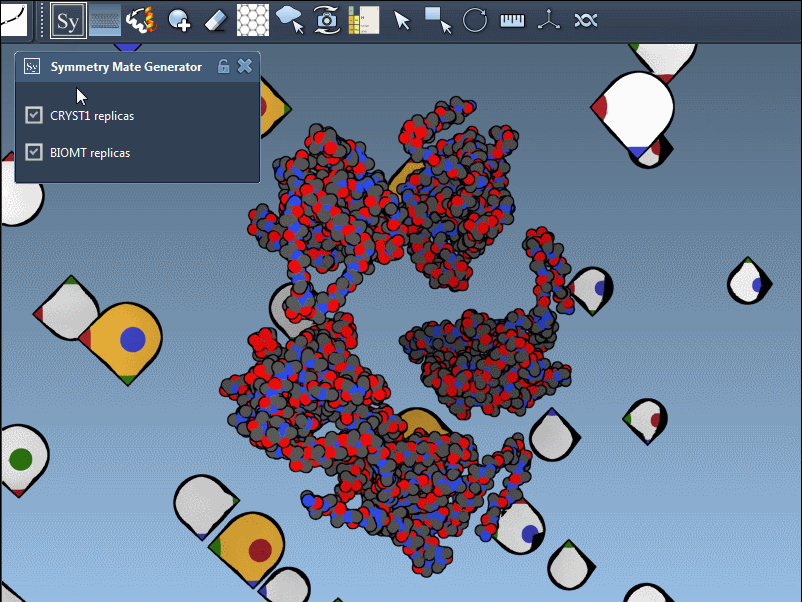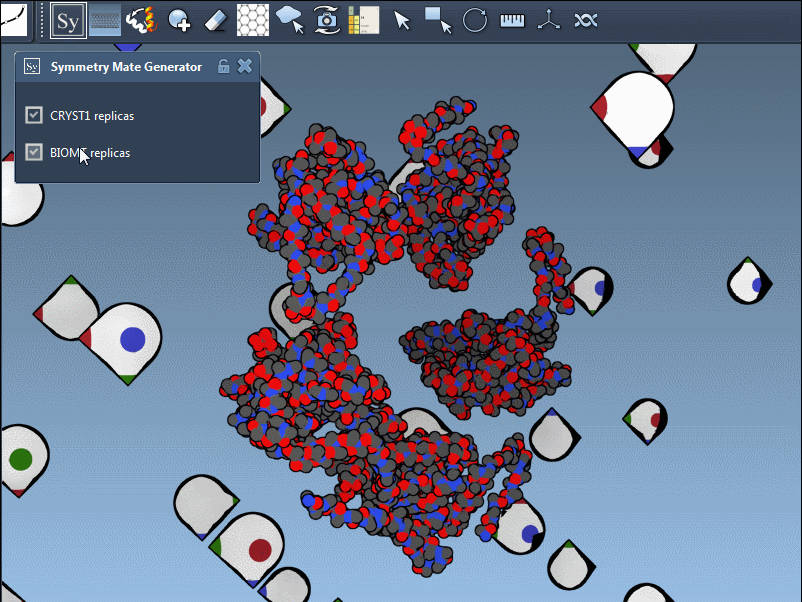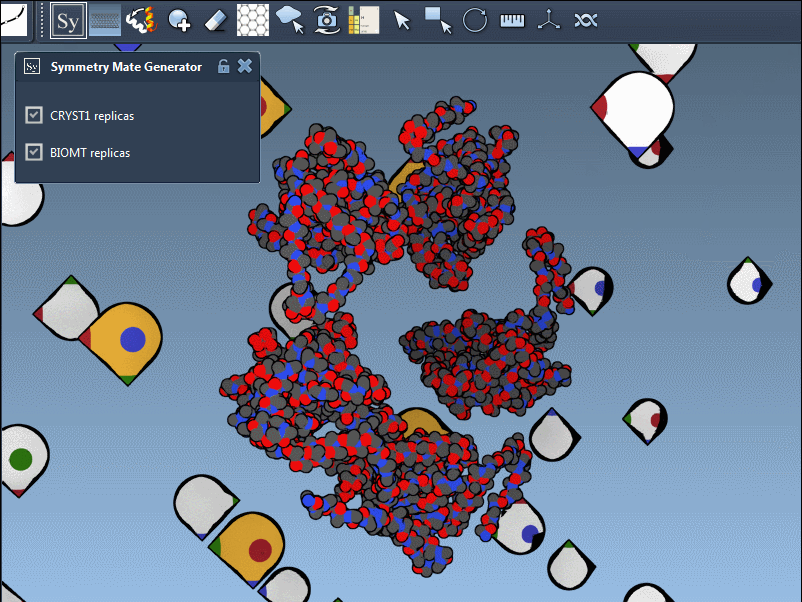
Once the extension is activated, SAMSON displays interactive control nodes in the 3D viewport. These represent symmetry transformations sourced from two types of PDB metadata: CRYST1 records (which describe the crystal lattice) and BIOMT records (which describe the biological assembly).
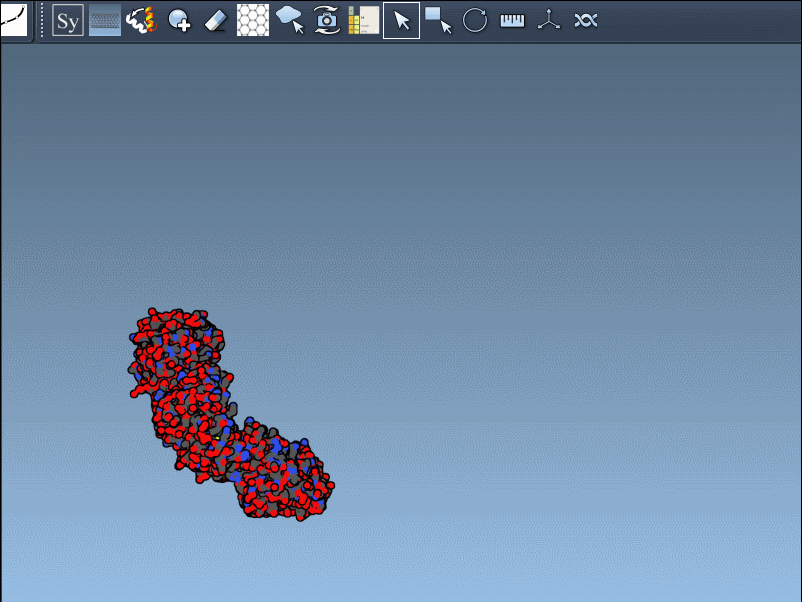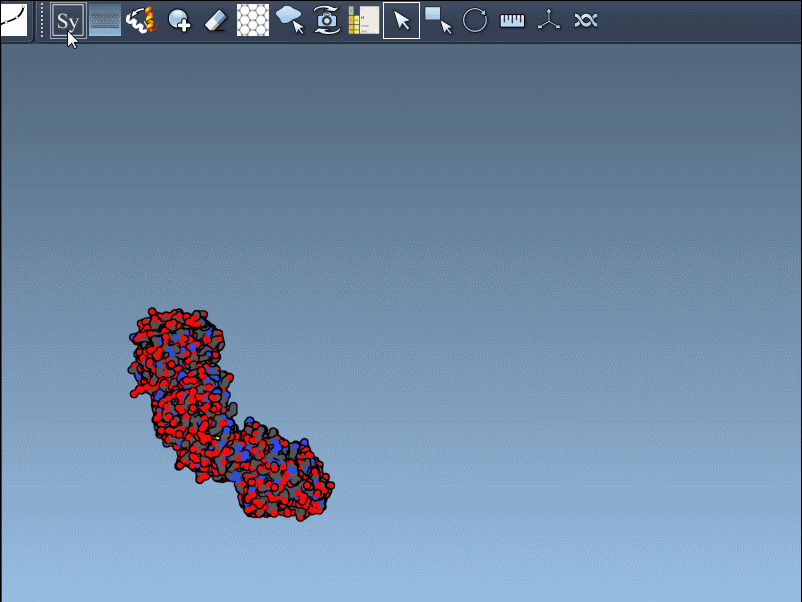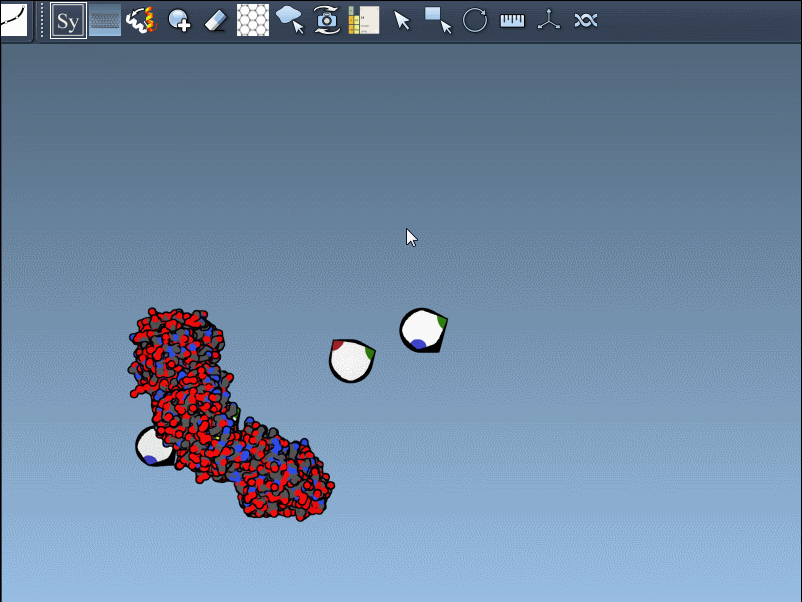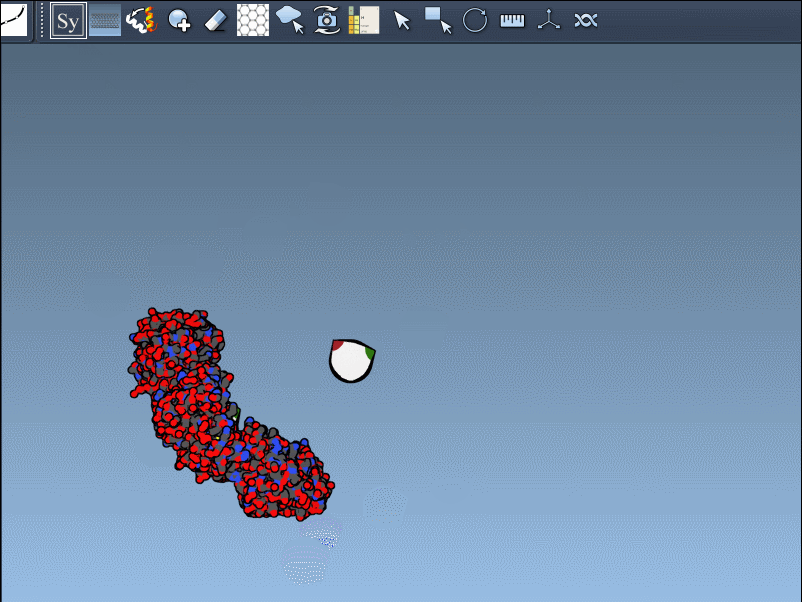
Hovering over a node previews the replica in real time. A simple click generates the actual symmetry mate, and you can undo it if needed using standard Ctrl/Cmd + Z. Hovering and clicking while holding Ctrl/Cmd allows you to generate all replicas related to that transformation in a single action, which is particularly useful when trying to view the complete quaternary structure at once.
Understand CRYST1 vs. BIOMT
Not all symmetry operations are the same. Some relate to how the molecule packs in the crystal lattice (CRYST1), while others show the biologically relevant organization (BIOMT). SAMSON color-codes these: white for CRYST1, yellow for BIOMT, helping you distinguish between the two visually.
Why This Matters
- Reconstructing full assemblies helps analyze protein-protein interfaces.
- You can identify symmetric binding sites relevant for drug discovery or design.
- Simulating the dynamic behavior of a multimer requires the full system.
- Designing oligomers or cages depends on symmetric arrangements.
Quick Usage Tips
- Use the mouse wheel while holding Ctrl/Cmd to adjust how many control nodes you see.
- Hover to preview, click to build, and undo when needed.
- Use the Ribbons visual model and color by chain to distinguish parts.
This interactive workflow significantly reduces prep time for downstream modeling and facilitates understanding of the symmetric nature of many biological structures. Whether you’re simulating interactions or designing supramolecular assemblies, having access to the full biological context is essential.
To explore step-by-step instructions and additional features, visit the official documentation page:
https://documentation.samson-connect.net/tutorials/symmetry/generating-symmetry-mates/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON here: https://www.samson-connect.net





