Any molecular modeler who’s ever tried to select a specific subset of atoms or residues in a large biomolecular structure knows the frustration: zooming in, clicking atom by atom, missing one, and starting over. But what if you could describe exactly what you want in a single smart sentence that the software understands immediately?
This is exactly what the Node Specification Language (NSL) in SAMSON offers. It lets you select atoms, residues, chains, or even abstract node groups using targeted expressions based on names, properties, topological relationships and more. Let’s take a closer look at how NSL can make your selections faster, more accurate, and less tedious.
What is NSL and Where Can You Use It?
NSL is a flexible language built into SAMSON that allows users to specify molecular selections using simple textual queries. You can use NSL in two places:
- The Find command (
Ctrl + F) - The Document view filter
In both places, NSL can replace manual clicking or dragging and help you find atoms meeting complex criteria—whether it’s selecting hydrogens, non-alanine residues, or carbons within a certain distance from a sulfur atom.
Say Goodbye to Manual Picking
Let’s say you want to select all backbone atoms that are part of alanine residues between residue IDs 20 and 50. You could write (short version):
|
1 |
n.t bb in r.t ALA and r.id 20:50 |
This expression tells SAMSON to find all nodes that are:
- Type:
backbone(short:bb) - That are part of residues type
ALA - Whose ID is between 20 and 50
This takes mere seconds, compared to minutes or more of manual work—and it’s reproducible.
Proximity Matters
NSL also supports spatial queries. Need all carbons within 5 angstroms of sulfur?
|
1 |
C within 5A of S |
Or in shorthand:
|
1 |
C w 5A of S |
This is perfect for analyzing binding pockets or hydrogen bonding patterns.
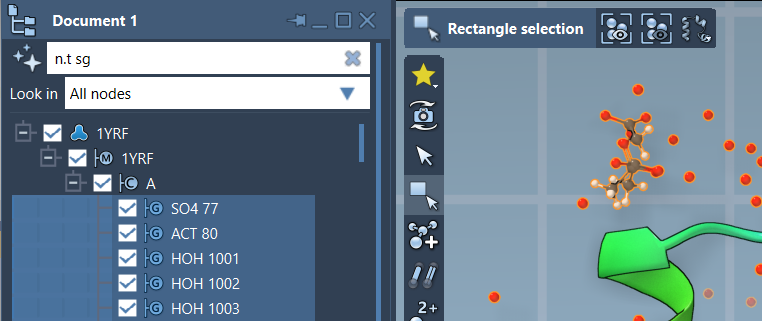
Using NSL in the Document View
In the Document view, NSL helps you filter nodes. For instance, to only display structural groups, write:
|
1 |
n.t sg |
Then just hit Enter to select them. This is often faster than navigating a deep document tree.
Let AI Help You Write NSL
If you’re not sure how to build your query, SAMSON includes an Ask AI button next to both the Find and Document view filters. The AI Assistant understands your open document and can generate the appropriate NSL for your goal—just describe what you want in plain language.
Takeaway
Using NSL can save you time, prevent selection errors, and help you focus more on analysis and less on interface navigation. Whether you’re dealing with massive protein complexes or small molecules, NSL enables more precise, flexible selections.
To learn more and explore more examples, visit the full documentation:
https://documentation.samson-connect.net/users/latest/nsl/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





