For molecular modelers and medicinal chemists, analog generation is often only half the battle. Once you’ve explored structural variations of a lead compound, the next big step is to evaluate how these analogs behave in 3D—especially if you’re preparing for docking simulations or interaction analysis.
But converting large sets of SMILES strings into reliable 3D models can be surprisingly tedious. Most workflows require external tools or scripts, often with inconsistent outcomes. What if you could go from a SMILES code to a chemically reasonable 3D structure—and do that directly inside your molecular modeling environment?
This is where the SMILES Manager extension in SAMSON offers a smooth, integrated experience. After generating analogs via positional analogue scanning, you can immediately convert any of the results into 3D structures for further analysis and simulations—without leaving the platform or needing additional software.
Why SMILES to 3D Matters
Working with molecules in 2D is handy for editing and visualization, but docking studies, conformational analysis, and molecular dynamics require accurate 3D geometries. Manually converting SMILES to 3D structures can become a bottleneck, especially as the number of analogs grows. Any error at this stage—like misplaced bonds or distorted geometries—can skew downstream results.
How It Works in SAMSON
Once you’ve run the positional analogue scanning process using the SMILES Manager, your analogs are listed in a table along with their 2D representations. From this point on, moving to 3D structures takes just one click:
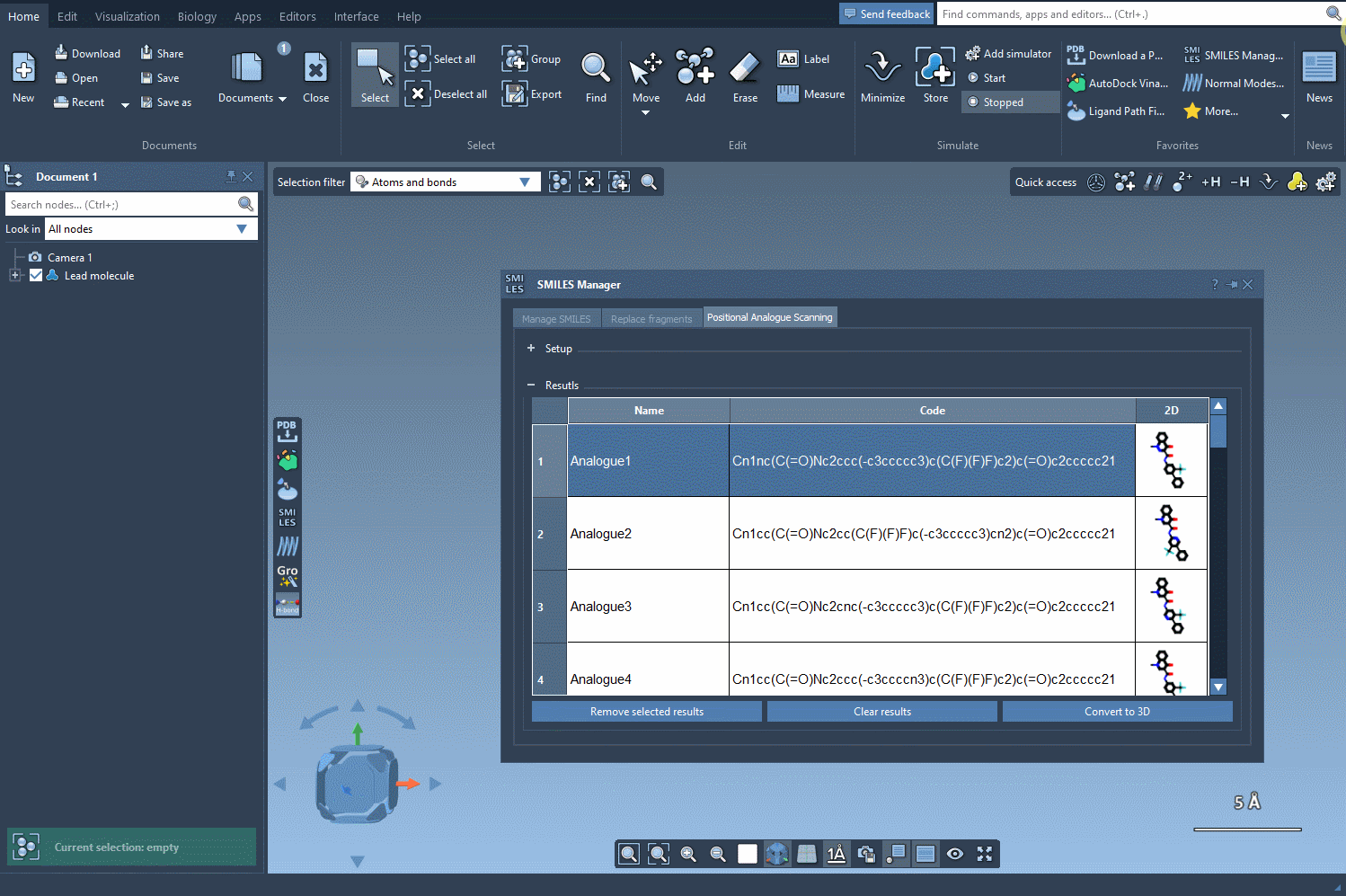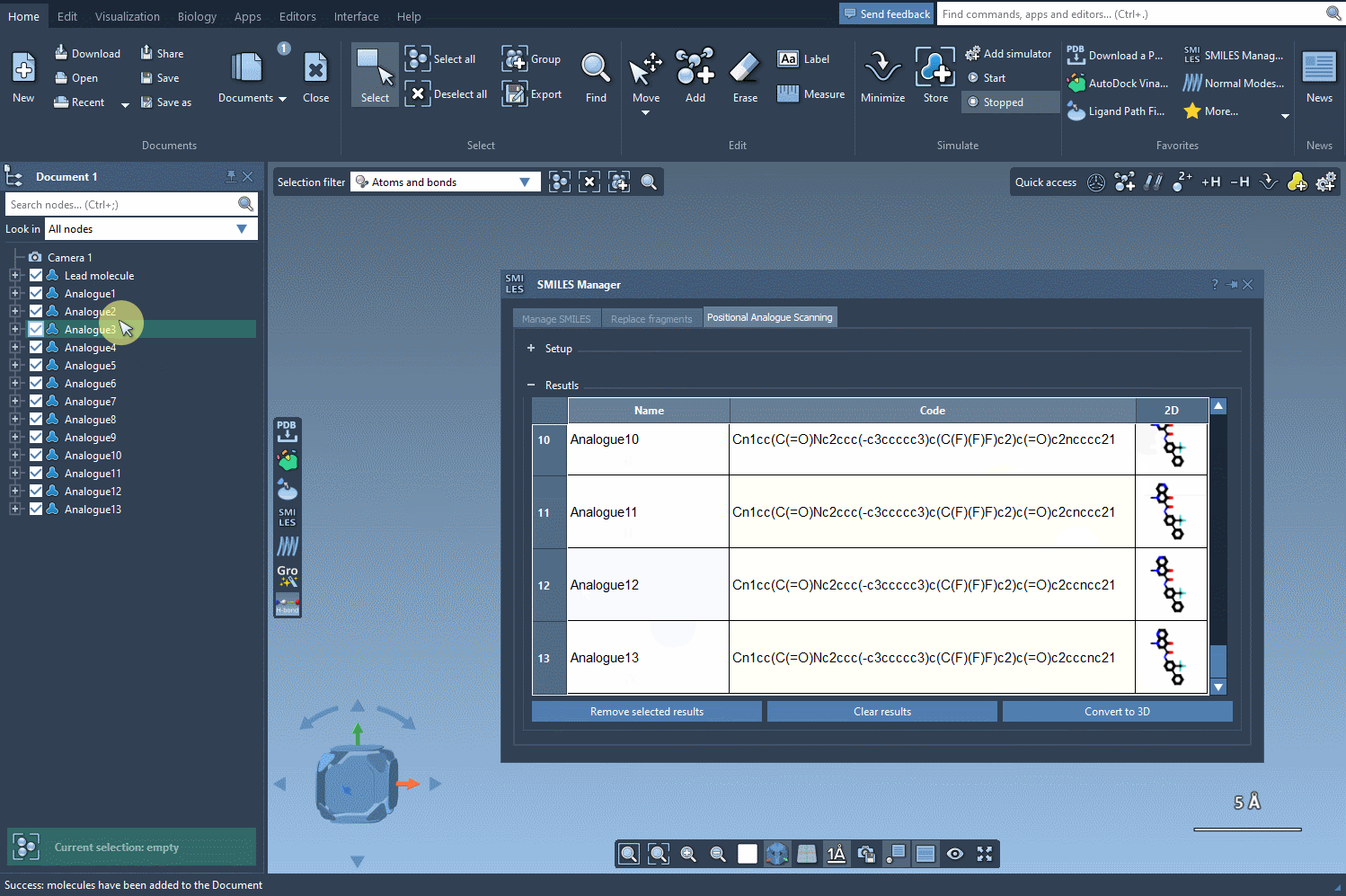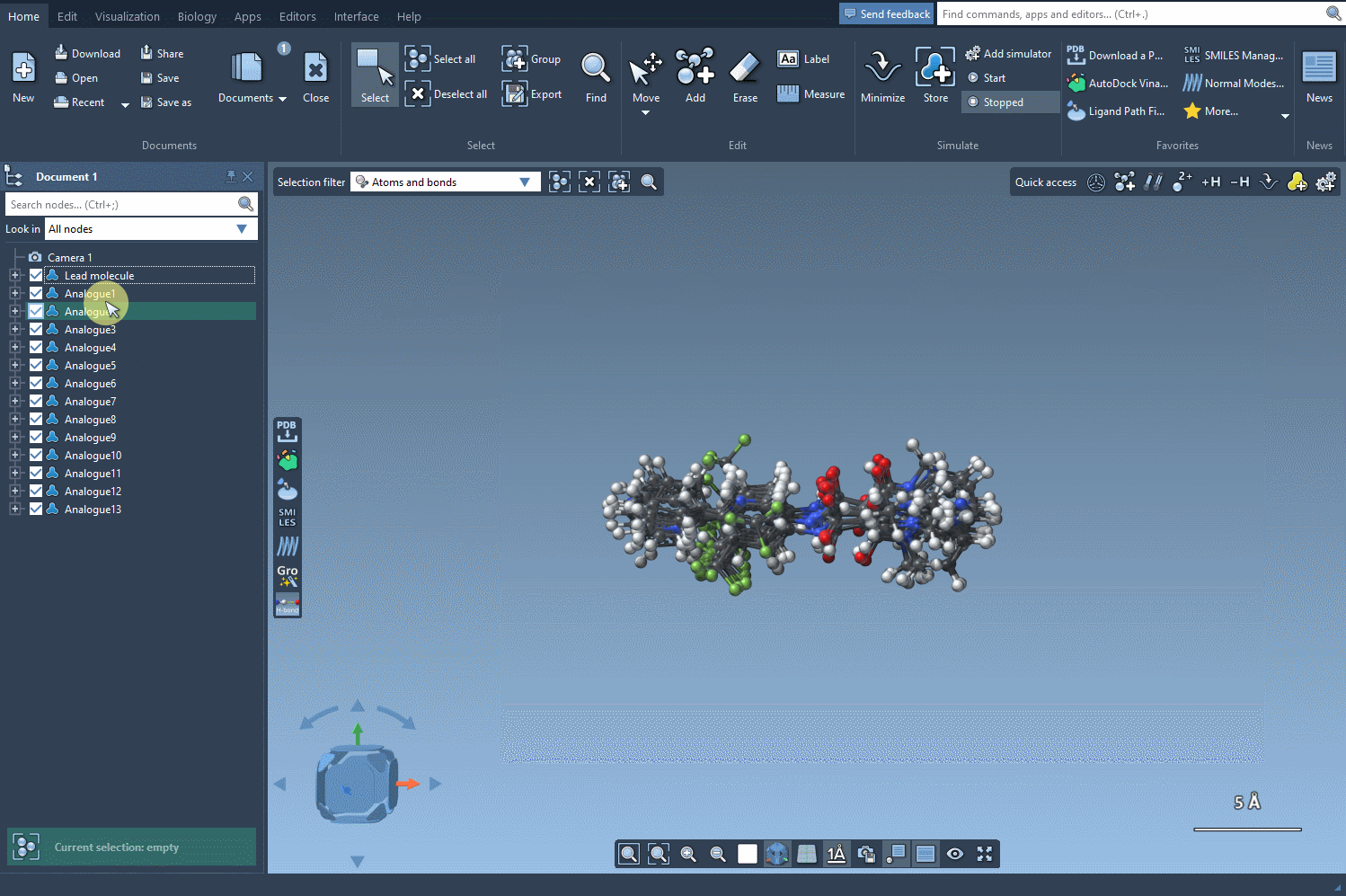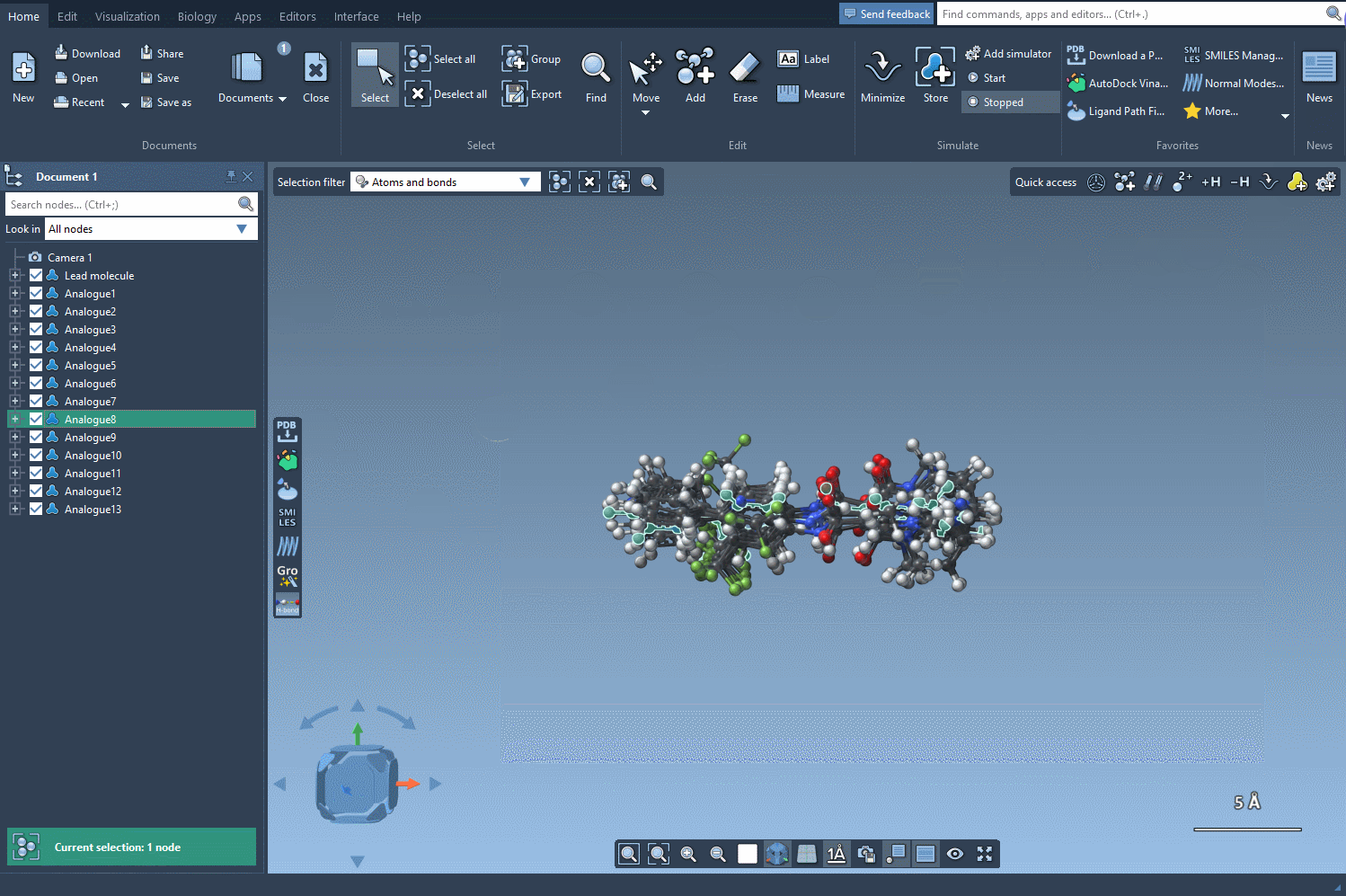
Click the Convert to 3D button, and SAMSON will generate 3D geometries for your selected analogs. The results are automatically placed in your SAMSON document, ready for docking (e.g., with the Autodock Vina Extended extension), electrostatics analysis, or simply for visual inspection.
A Closer Look at the Process
This step isn’t just cosmetic. The generated 3D structures are chemically meaningful, suited for immediate use. You don’t have to worry about broken rings, incorrect stereochemistry, or starting structures that need heavy tweaking before minimization or simulation. Each structure is also context-aware: built from the original SMILES code and taking into account bonding patterns during rendering.
From Prototype to Simulations
Once 3D structures are available, you can begin answering key questions:
- Do specific substitutions preserve or disrupt binding modes?
- Are new compounds making favorable contacts with the target protein?
- How does a methyl vs. fluorine group shift the geometry of the active core?
The seamless transition from 2D to 3D makes it fast and convenient to test these hypotheses. All the analogs you generate can be aligned, docked, and visually inspected directly in SAMSON, enabling a more focused iteration loop in your design process.
To learn more about the entire workflow—starting from SMILES input all the way to analyzing interactions—visit the full documentation for positional analogue scanning in SAMSON: View Full Documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. Get started at https://www.samson-connect.net.





