Preparing realistic molecular environments around membrane proteins can be a time-consuming task. Building lipid bilayers manually, aligning structures, avoiding bad contacts — this setup work often becomes the bottleneck before simulation even begins. If you’re using SAMSON, there’s a dedicated tool that may simplify this process significantly: the Molecular Box Builder.
This blog post walks you through using SAMSON’s Molecular Box Builder to rapidly create lipid layers—or full bilayers—around proteins, making it easier to focus on what matters: analysis and simulation.
Why this matters 🔍
Accurate modeling of membrane proteins often requires embedding them correctly in a lipid environment. Poor alignment, overlapping atoms, or inaccurately sized boxes can lead to simulation artifacts or computational errors. Traditional tools can involve a significant learning curve or scripting. By contrast, Molecular Box Builder combines a visual interface with precise control over placement and orientation of molecules.
Step-by-step: Building a lipid layer
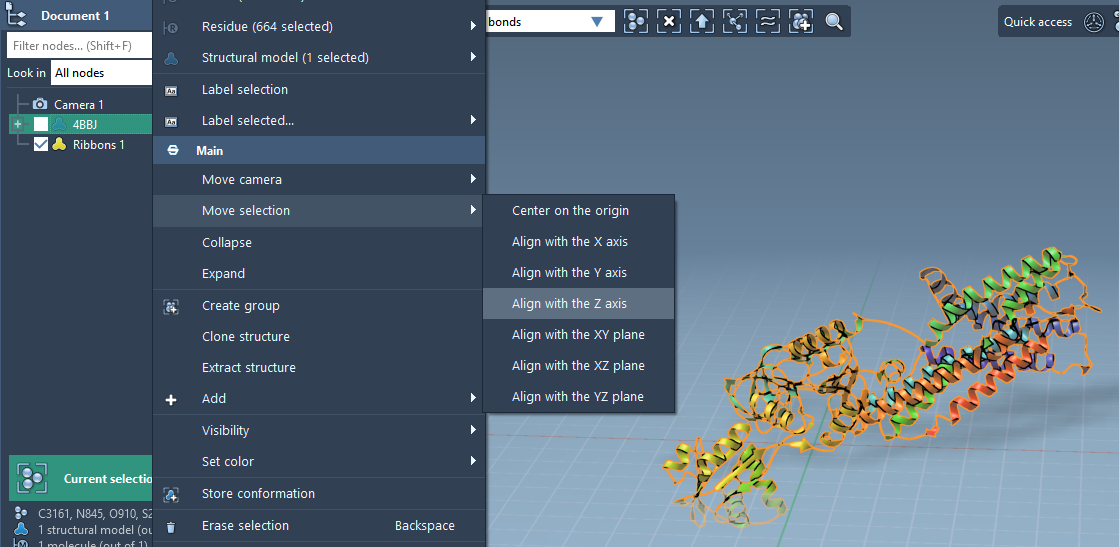
1. Orient your protein
To build the membrane accurately, orient the protein so that the membrane aligns with the Z-axis. In SAMSON:
- Right-click the protein in the Document view.
- Select Move selection > Align with Z axis.
- Then choose Move selection > Center on the origin.
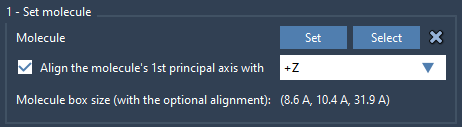
2. Set your lipid molecule
- Import a lipid molecule of your choice.
- Select it and click Set in Molecular Box Builder.
- Align its principal axis to
+Zto match orientation.
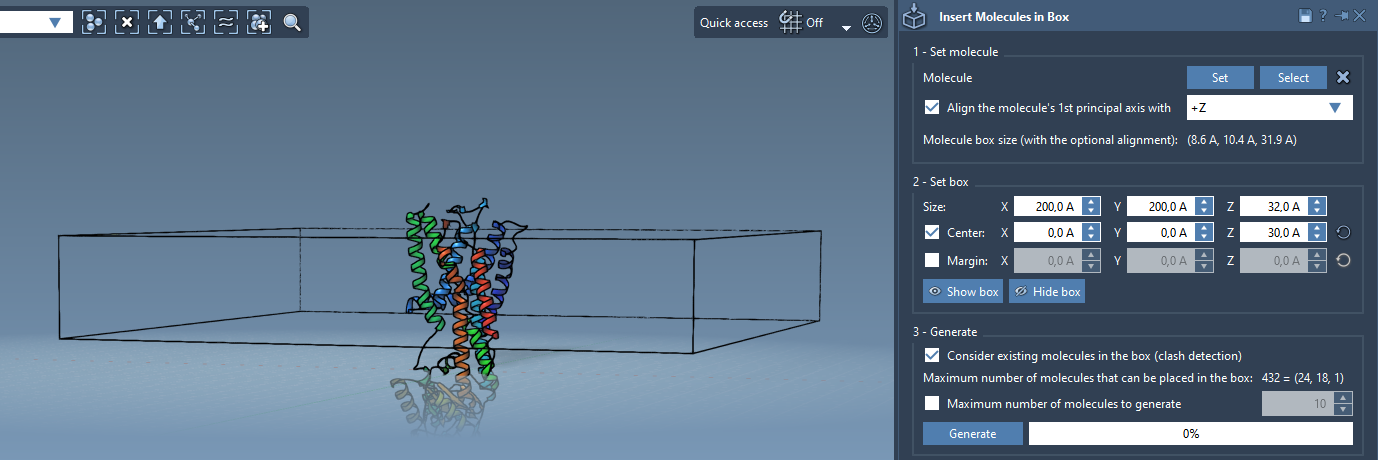
3. Define the box
Now, center the box around the protein to define the lipid layer’s bounds. Adjust the box size depending on the thickness and lateral dimensions of the desired layer. Set appropriate margins to prevent overlap or insert slightly compressed layers for high-density packing.
4. Generate the lipid layer
- Enable Consider existing molecules in the box to avoid interfering with the existing protein structure.
- Click Generate. The lipids will populate the box while avoiding the protein volume.

Building a full bilayer
To go beyond a single leaflet and create a bilayer:
- Generate the first leaflet with the lipid aligned to
+Z. - Shift the box downward so the next layer stacks below.
- Generate the second leaflet with the lipid aligned to
-Z.
This symmetrical placement ensures correct membrane topology and can be the basis for further solvation and ion placement (using additional tools like PDBFixer).
What’s next?
Once your bilayer is ready, you can minimize and simulate the system using SAMSON’s GROMACS Wizard. This integrated workflow reduces setup time significantly while allowing for full visual validation of every step.
To learn more, consult the Molecular Box Builder documentation.
*Note: SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





