Modeling carbon nanotubes (CNTs) is a common yet often tedious task in molecular design and materials science. Whether used for simulations in electronic properties, nanodevices, or biomolecular systems, building precise CNT structures can be surprisingly error-prone and time-consuming—especially if done from scratch with general-purpose modeling tools.
If you’ve ever struggled to adjust chiral indices or found yourself buried in command-line generators for carbon nanotubes, SAMSON offers an easier way. With the Nanotube Creator Extension, you can build single-walled and multi-walled CNTs interactively using your mouse. No more trial-and-error with parameters—you see what you build, in real-time.
Build What You Need, Visually
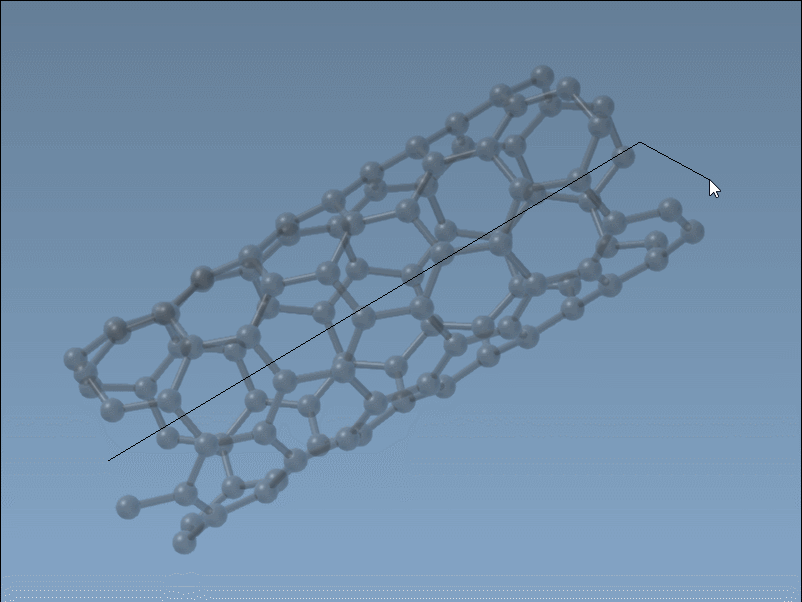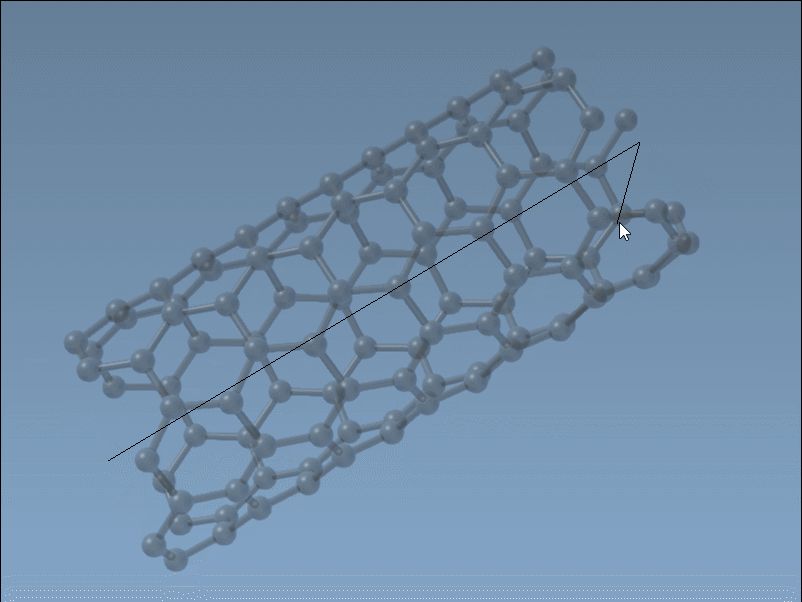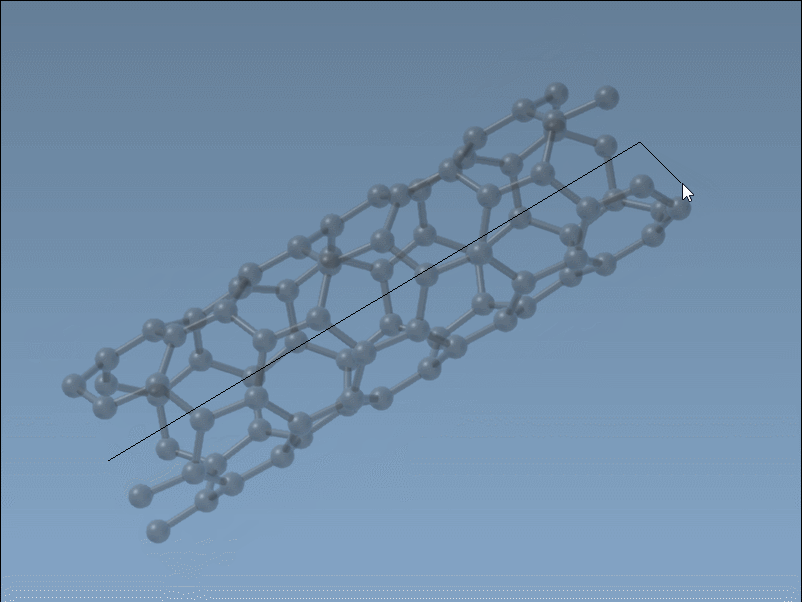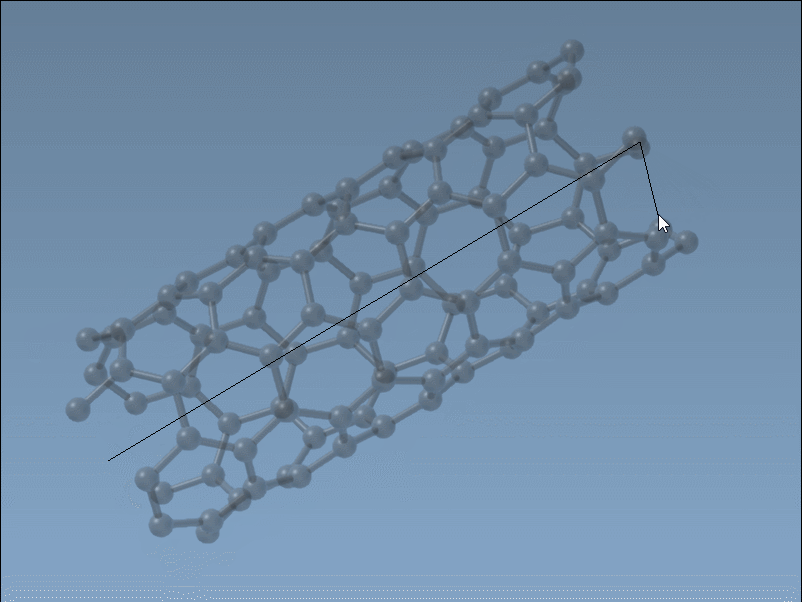
The Nanotube Creator in SAMSON introduces an editor that lets you construct CNTs in two simple mouse-driven steps directly in the viewport:
- Set the axis and length by pressing and dragging the left mouse button. This step defines the direction and number of unit cells (the
nparameter). - Adjust the radius by moving your mouse and clicking again. This sets the
mparameter.
While building, live feedback appears in the status bar, showing you the precise chiral indices you’re selecting. This is ideal when you want to generate specific structures (armchair, zigzag, or chiral), or simply test variations visually without recalculating coordinates.
Fine-Tuning with a GUI
For those who prefer full control over numerical values, the same extension also includes a graphical interface (GUI). It lets you manually enter parameters like n, m, and 3D positions of the tube ends. The GUI is especially effective for creating multi-walled CNTs or for replicating structures from literature with precision.
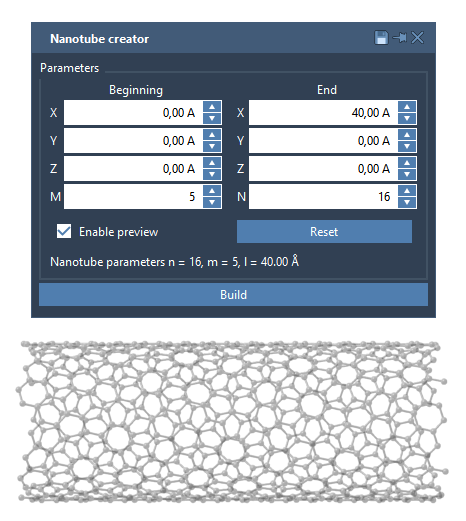
A Quick Example: Multi-Walled Nanotubes
Thanks to the GUI and real-time visualization, you can create concentric nanotube structures for advanced materials modeling or molecular transport. For example, try this 3-layer multi-walled setup:
- Start/end positions:
(0, 0, 0)to(40, 0, 0). - Layer 1:
n = 6,m = 6 - Layer 2:
n = 10,m = 10 - Layer 3:
n = 14,m = 14
Click Build after each layer to assemble your structure. This approach lets you design full nanotube-based systems for molecular simulations, sensors, nanopores, and more.

Takeaways for Molecular Modelers
If you frequently explore CNT properties or integrate nanotubes in nanoscale simulation workflows, using SAMSON’s interactive tools significantly reduces the friction. Instead of generating coordinates externally and importing files, you build, adjust, and reconfigure directly in your workspace—matching the way scientists actually think and iterate.
Learn more and follow the full tutorial here: Building Carbon Nanotube Models in SAMSON.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





