For many molecular modeling tasks, especially those involving large biological assemblies like viral capsids or protein complexes, understanding molecular symmetry can save time and reduce complexity. But automatic symmetry detection isn’t perfect—sometimes, you already know the answer, and it’s best to tell the software directly. If you’ve ever had a model where you know the expected symmetry group (say, D3 for a trimeric protein), SAMSON’s Symmetry Detection extension lets you specify it manually.
Here’s why that can be helpful, and how to do it.
Why Specify Symmetry Manually?
When working with large molecular assemblies, automatic symmetry detection may propose multiple plausible symmetry groups. While this feature is useful for exploration, it can introduce ambiguity when you already have experimental data or prior knowledge about the system’s symmetry.
For example, if you’ve looked up a structure’s symmetry in the PDB or scientific literature, you might expect a specific symmetry type—such as D3 for the structure 1B4B. Rather than reviewing multiple candidates, you can go straight to the result you trust and focus on what matters most: analyzing interfaces, designing mutations, or setting up simulations for the asymmetric unit.
How to Set Symmetry Manually in SAMSON
- Launch SAMSON, and load your biological assembly (e.g.,
1B4B). - Open the Symmetry Detection extension from Home > Apps > Biology.
- Click Compute symmetry to initialize the analysis.
- If you know your expected symmetry type, select it directly from the dropdown menus. For example, set the group to
D3for a dihedral group of order 3. - The extension will then display the corresponding axes, enabling you to align, visualize, and interact with them just like with automatically detected ones.
Use Cases and Benefits
- Focused Validation: If you’re testing a hypothesis or validating an experimental structure, using known symmetry helps confirm structural correctness without distraction from alternative options.
- Simulation Preparation: Defining symmetry upfront makes it easier to isolate the asymmetric unit needed for simulations, avoiding redundancy.
- Educational Use: Teaching about symmetry is more straightforward with clear, controlled examples using expected results.
Visual Feedback
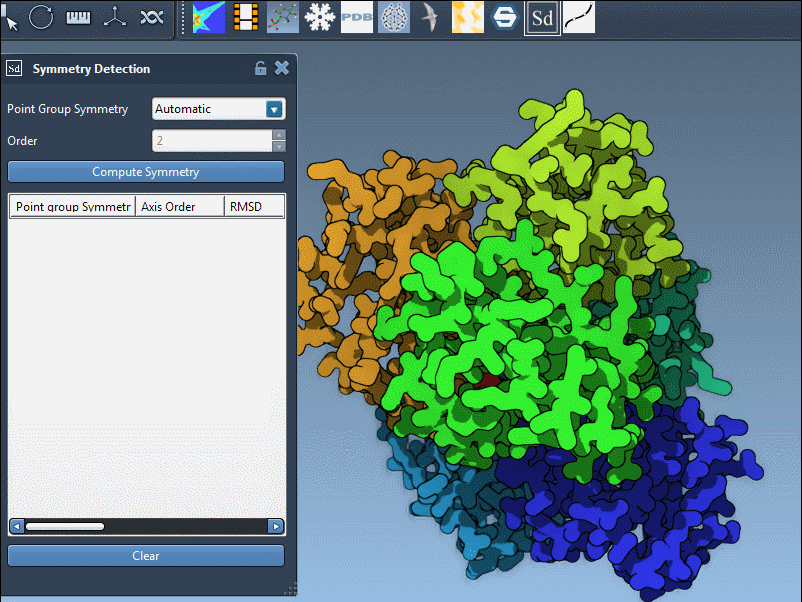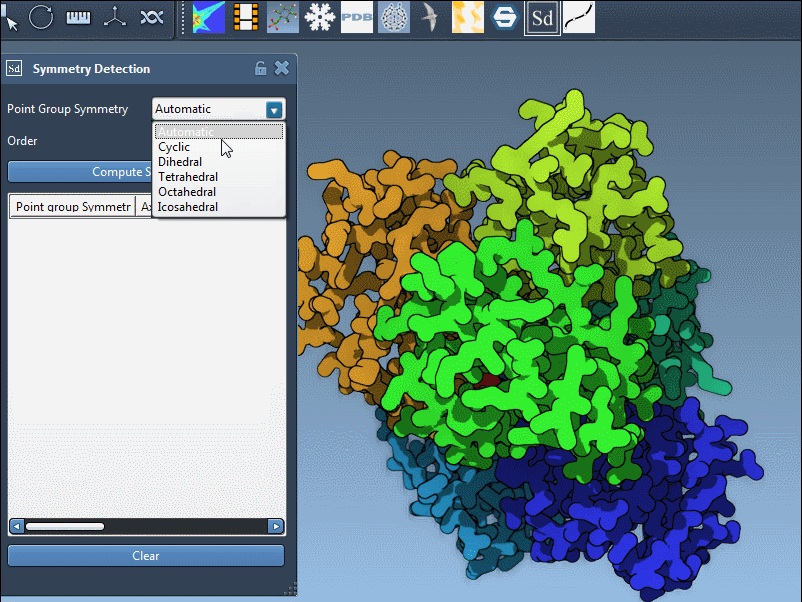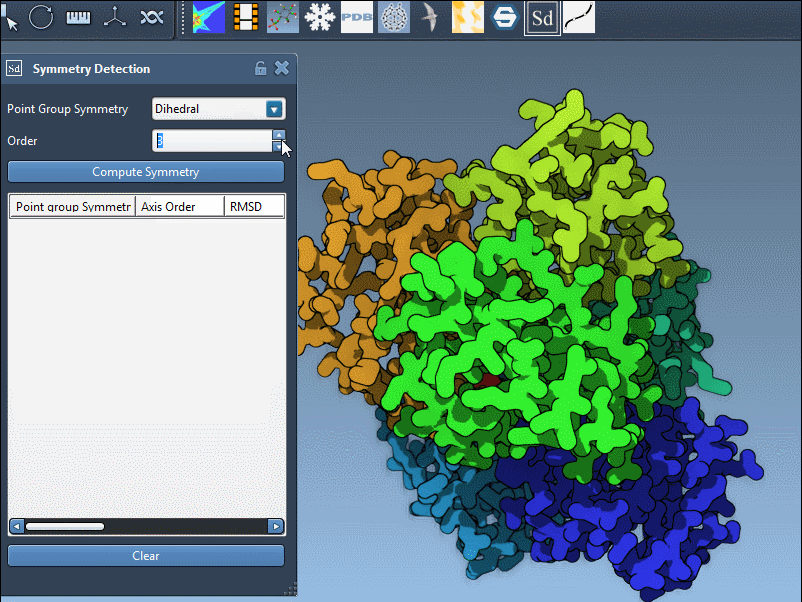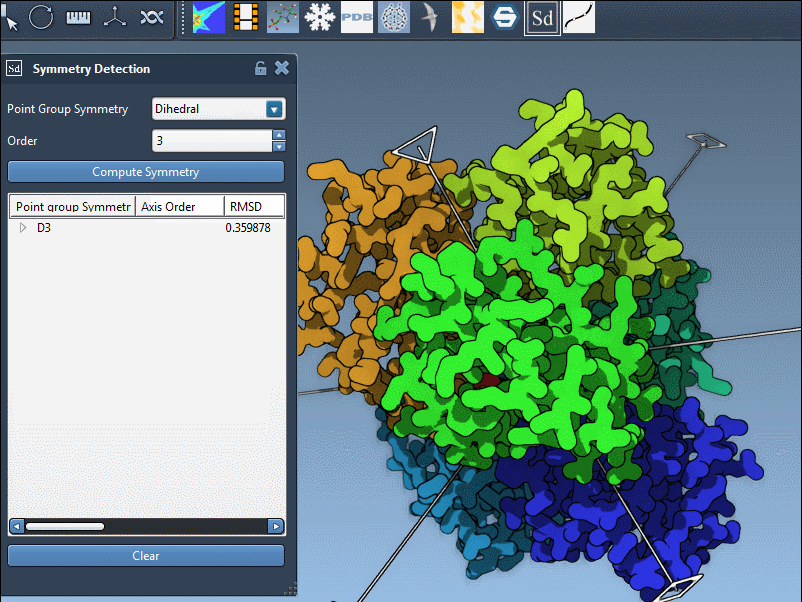
Once specified, symmetry axes appear in the viewport. For example, selecting D3 for 1B4B reveals a set of axes that you can interact with. Single-click to highlight an axis, or double-click to align the view for a better structural perspective.
Final Tip
Even when specifying symmetry manually, it’s helpful to review per-axis RMSD values to confirm that the structure closely matches the expected ideal. This hybrid approach—personal knowledge plus computational feedback—improves confidence when designing or validating molecular systems.
To learn more about symmetry detection and its applications in SAMSON, visit the full documentation page here.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





