When modeling protein structures, especially those involved in viral entry mechanisms, capturing their motion from one functional state to another is often challenging. For molecular modelers investigating SARS-CoV-2 or designing therapeutics targeting protein conformations, having access to a visual and modifiable trajectory between spike protein states can be a significant advantage.
In this post, we highlight how SAMSON, the integrative molecular design platform, enables you to generate and visualize the conformational change of the SARS-CoV-2 spike protein—from the closed (down) state to the open (up) state—using interpolation and post-processing techniques for trajectory construction.
Why simulate spike motion?
Understanding the spike protein’s opening mechanism is critical because it reveals how the virus binds to the ACE2 receptor on human cells. This structural shift, from a closed to an open state, is central to entry and infection. Furthermore, it is an attractive target for neutralizing antibodies and therapeutic inhibitors.
Experimental methods offer us two main structures: PDB 6VXX (closed state) and 6VYB (open state). But how do we model the motion between them convincingly, especially when the structures don’t share the exact same number of residues?
Interpolation + P-NEB = Motion
SAMSON enables the computation of plausible transitions through a combination of its As-Rigid-As-Possible (ARAP) Interpolation Path module and the Parallel Nudged Elastic Band (P-NEB) module. Here’s how the trajectory was created:
- Hydrogens were added and structures minimized for 6VXX and 6VYB, adjusting for sugar residues.
- ARAP was used to interpolate an initial path from the open (6VYB) to the closed state (6VXX).
- Since the structures differ in residue count, a modified version of the closed structure was extracted from the ARAP path to better align with the open-state structure.
- A second interpolation was then performed using this adjusted structure.
- The path was refined using P-NEB to ensure a smoother, more realistic transition.
Visualizing the transition
Below are animated views showing the spike movement between the closed and open conformations. These perspectives help not only in understanding how ACE2 recognition is enabled but also in locating druggable conformational intermediates.
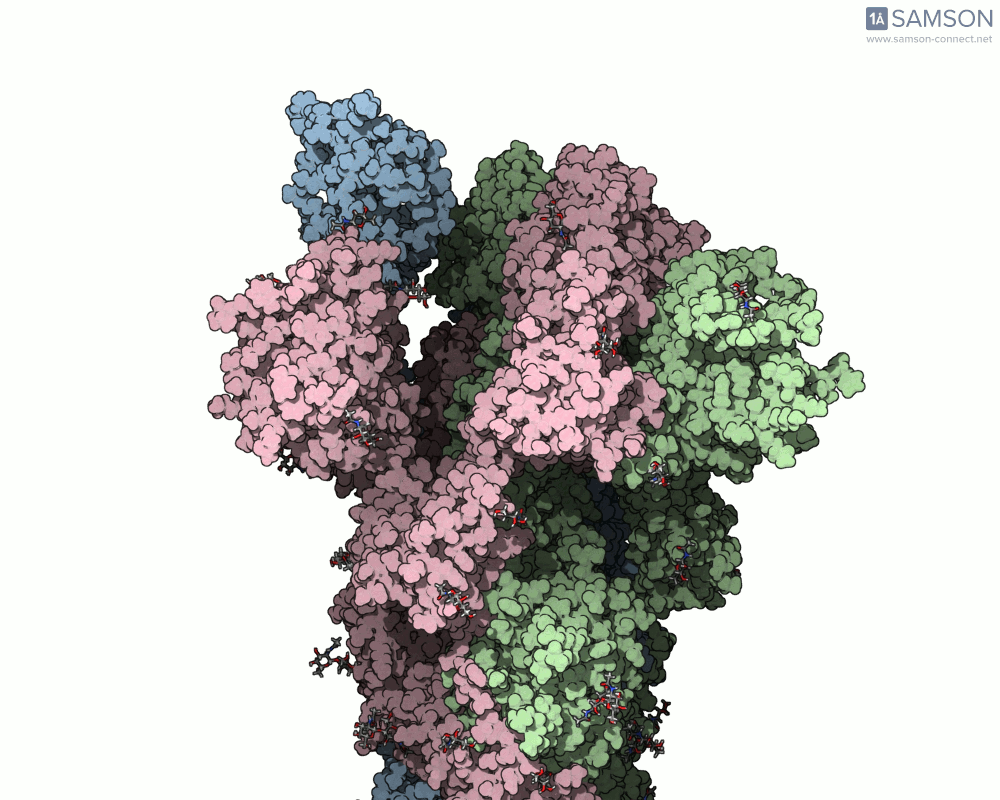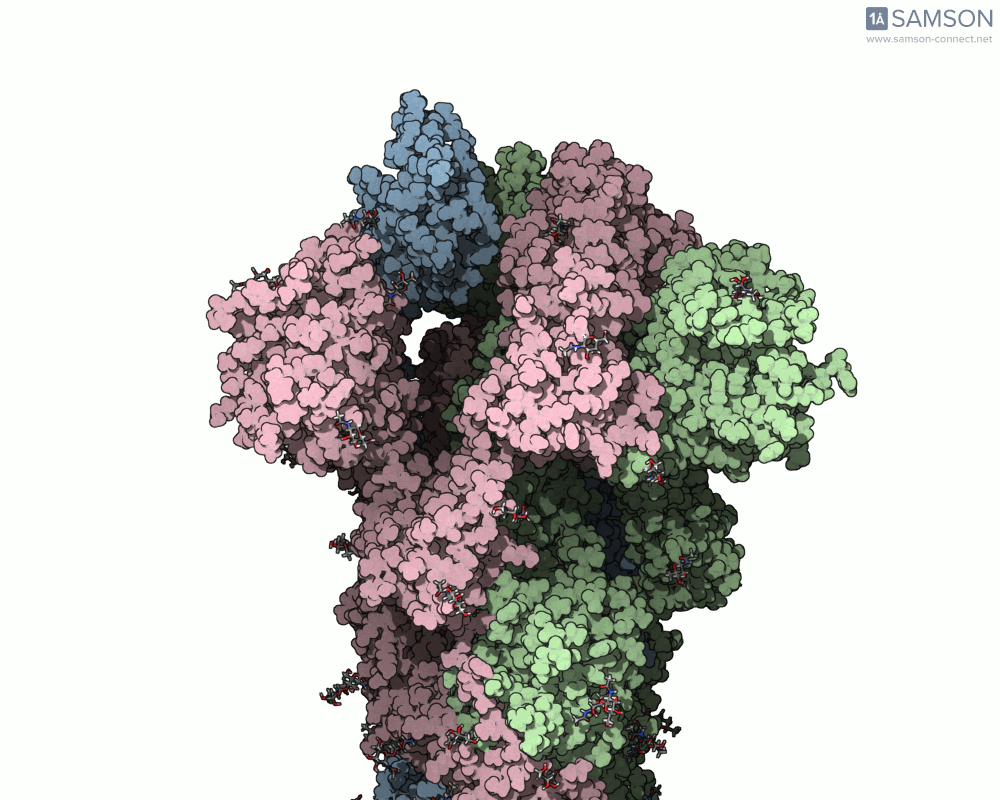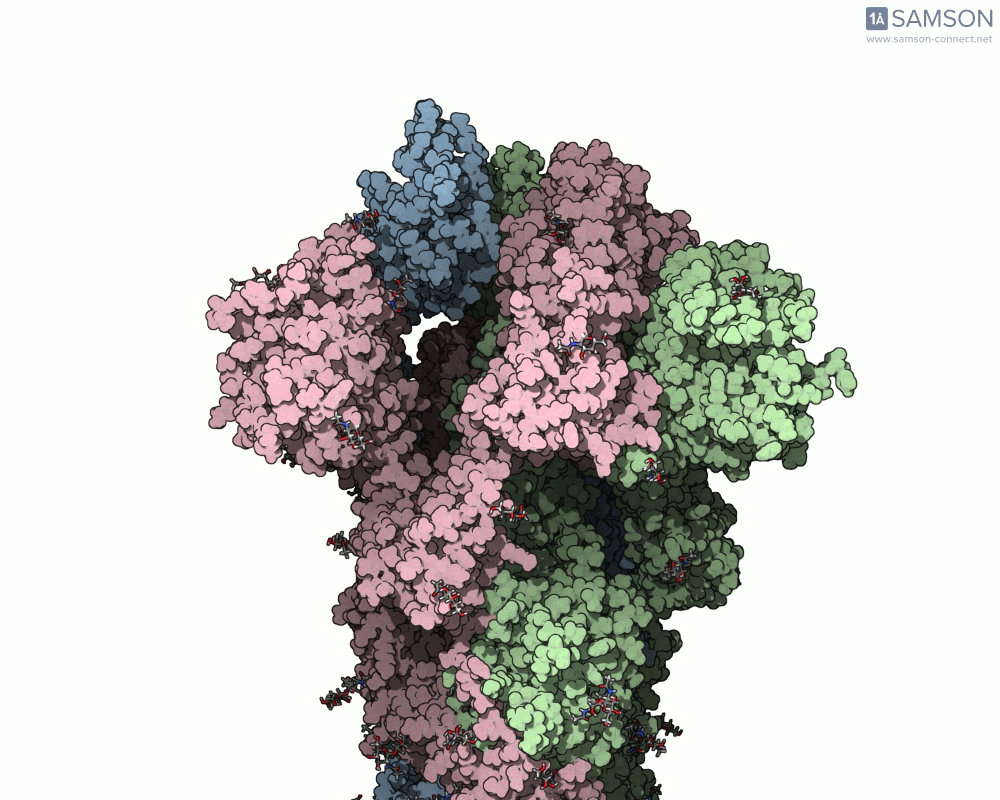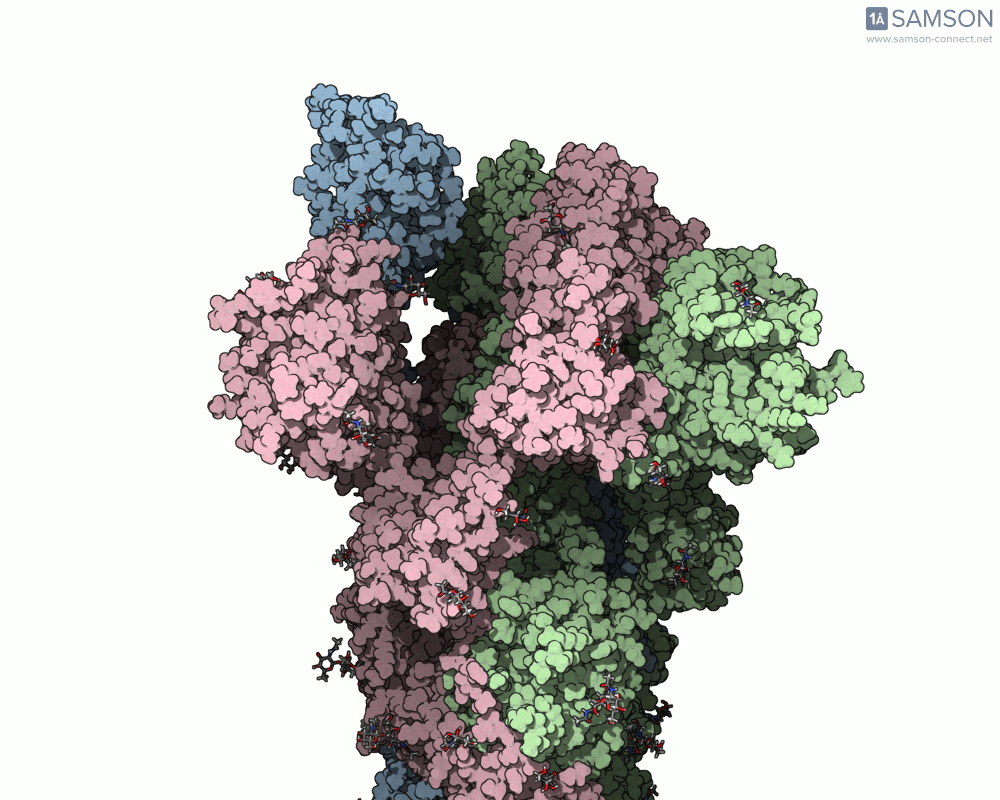
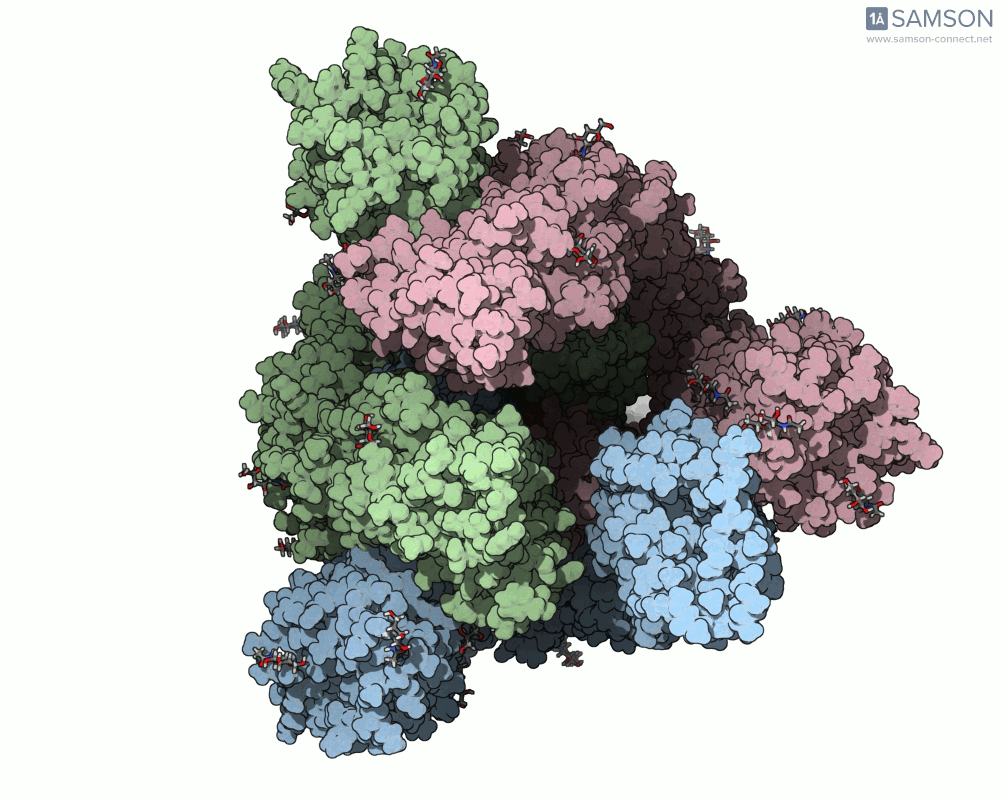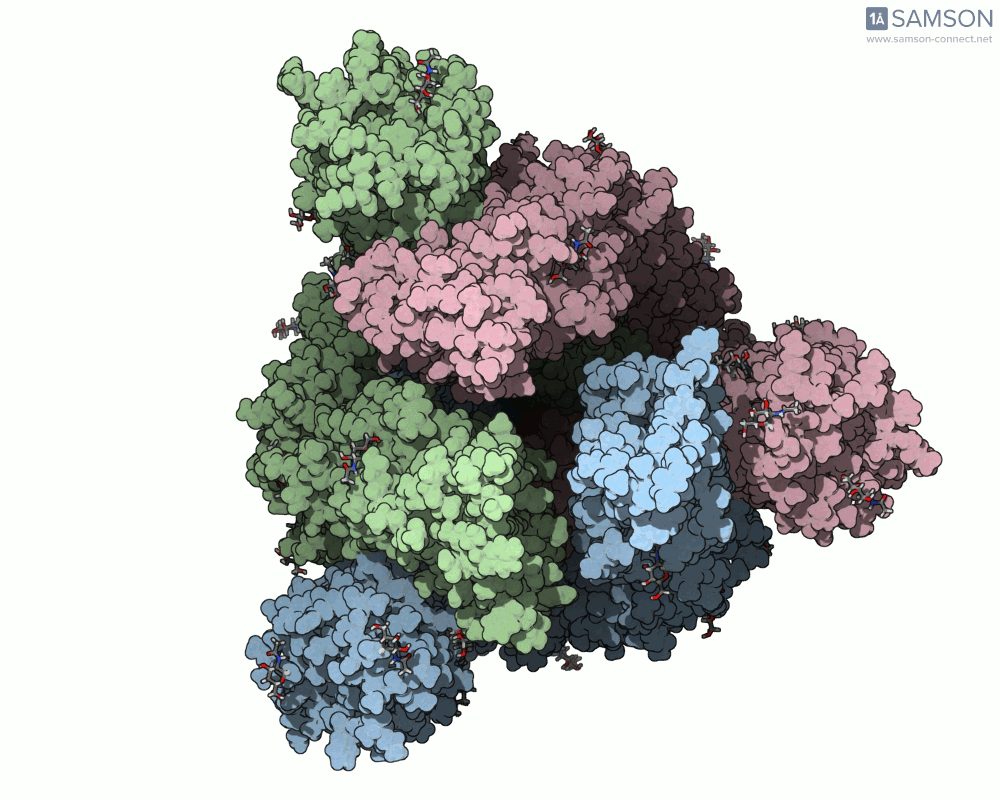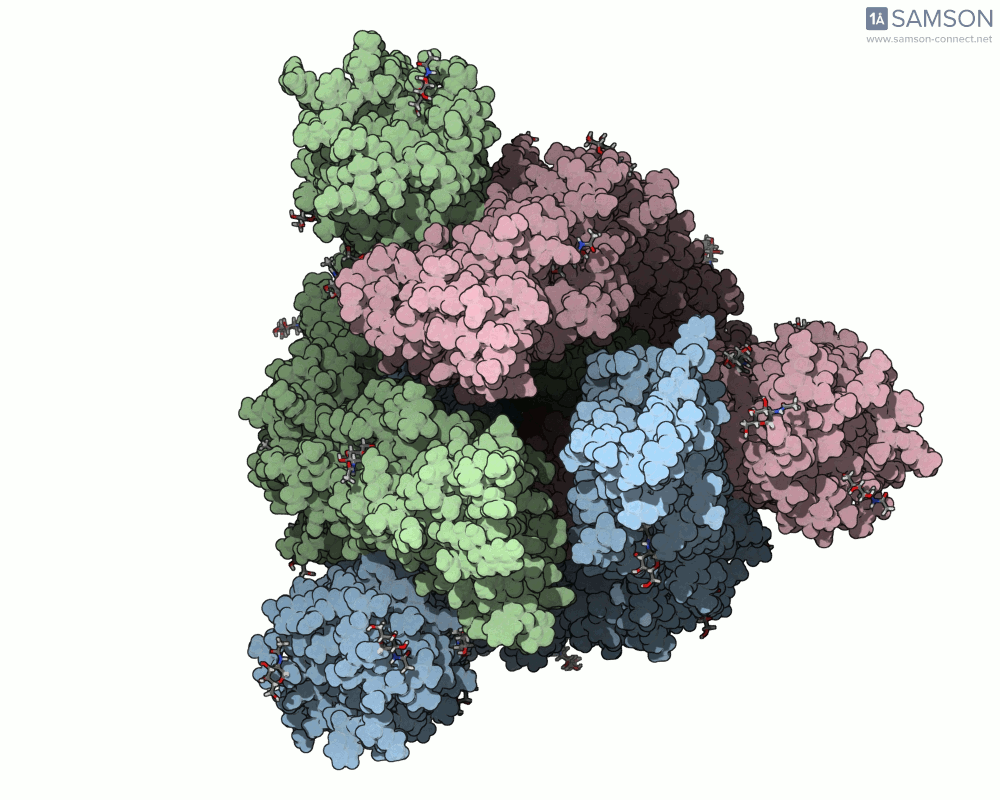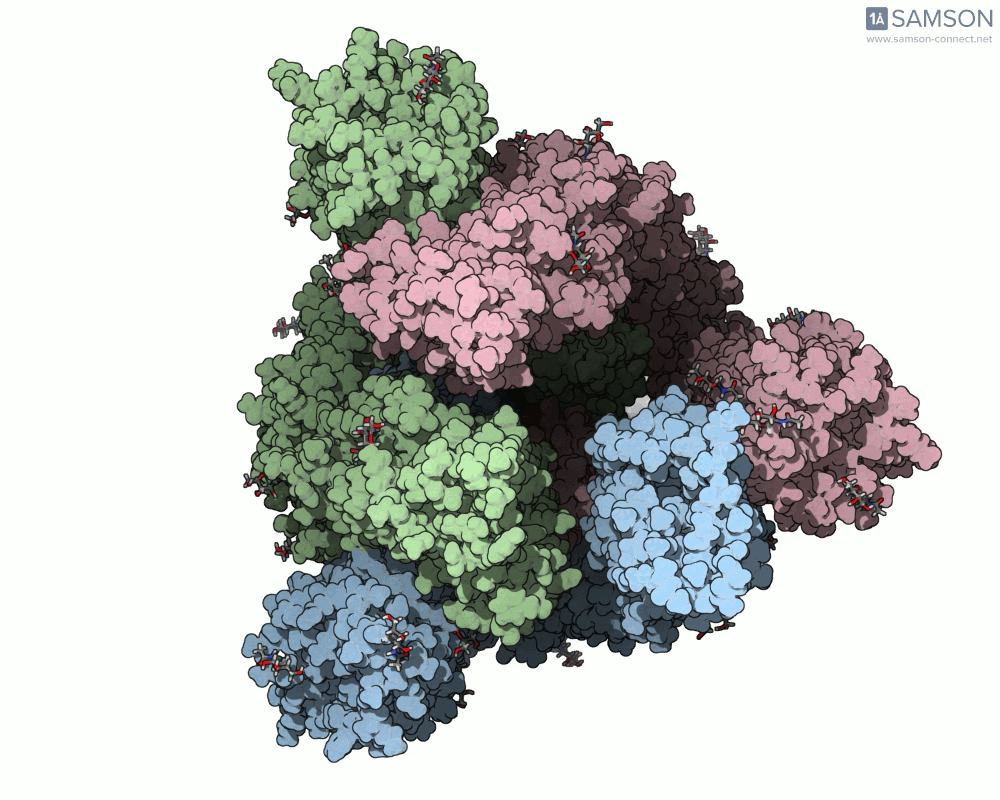
Get the trajectory files
To dig deeper or use these motions in your own analysis, you can download the trajectory as:
The SAMSON version is especially useful, integrating the animation path and structure with interactive viewing and playback.
Good to know
Note that these trajectories are illustrative and computational—they have not been experimentally validated. However, they can be useful for further simulations, visualizations, or hypothesis generation in drug discovery efforts.
For more details, read the full tutorial on SAMSON’s official documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





