When you’re working with molecular design tools, one of the most frequent and sometimes frustrating tasks is setting up your initial molecule just to start with a new technique or analysis. Whether you’re exploring new analogs or experimenting with substitutions, having to retype or re-import molecular structures repeatedly can slow you down and lead to small errors early in your workflow.
If you’re using SAMSON, there’s a convenient shortcut when working with the SMILES Manager: you can skip manual SMILES entry and simply start with a molecule that’s already in your SAMSON document.
Making Use of What You Have
Here’s the situation: you already have a molecule visualized in SAMSON, maybe one you’ve downloaded from a database, built manually, or imported from another application. Rather than grabbing the SMILES string separately — or worse, searching for it online — just use what’s already loaded.
To do this, open the SMILES Manager and click the Use selection button. The selected molecule becomes your starting point automatically. This is especially useful if you’re screening a series of similar compounds or testing analogs of a known active molecule.
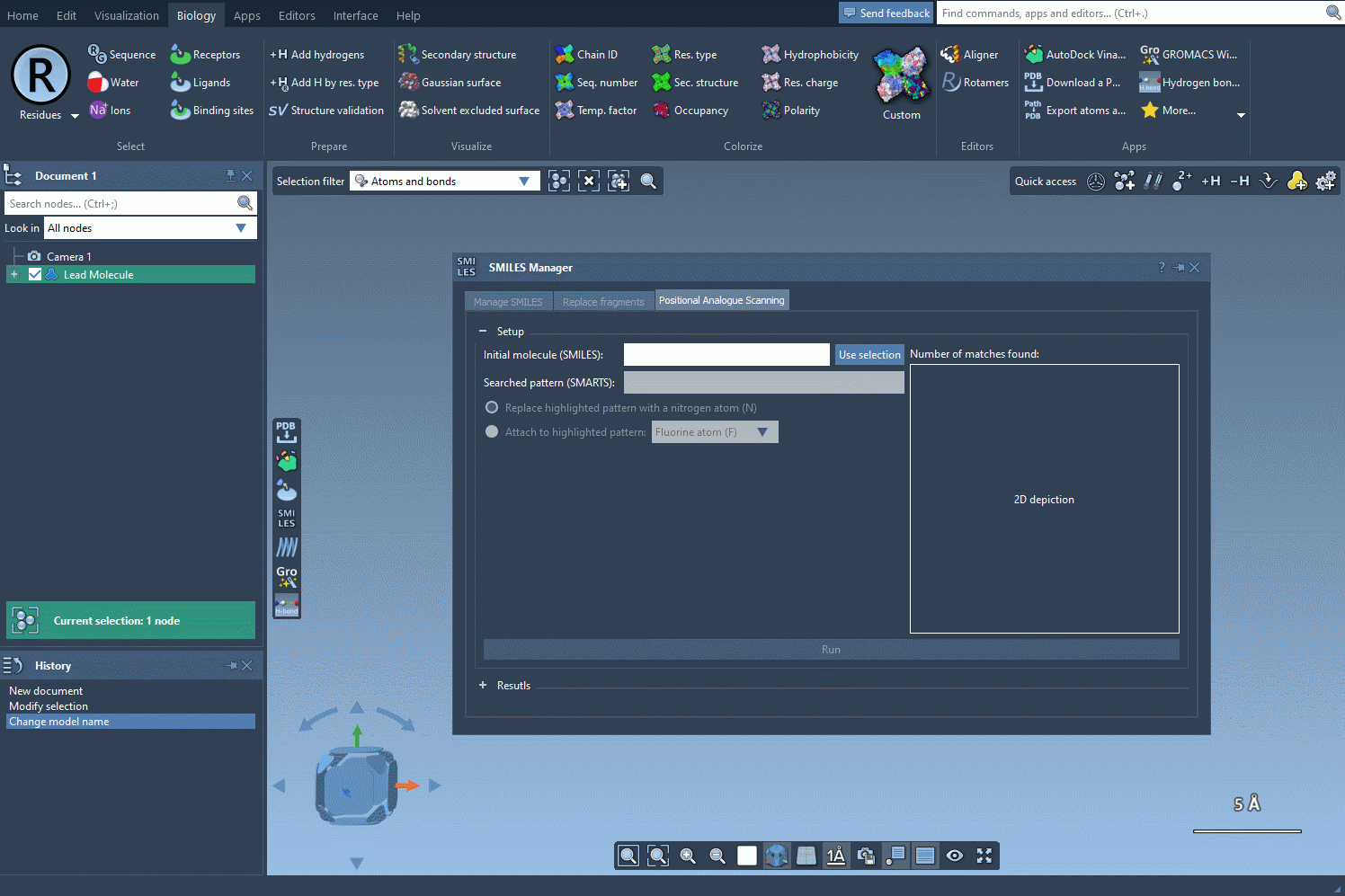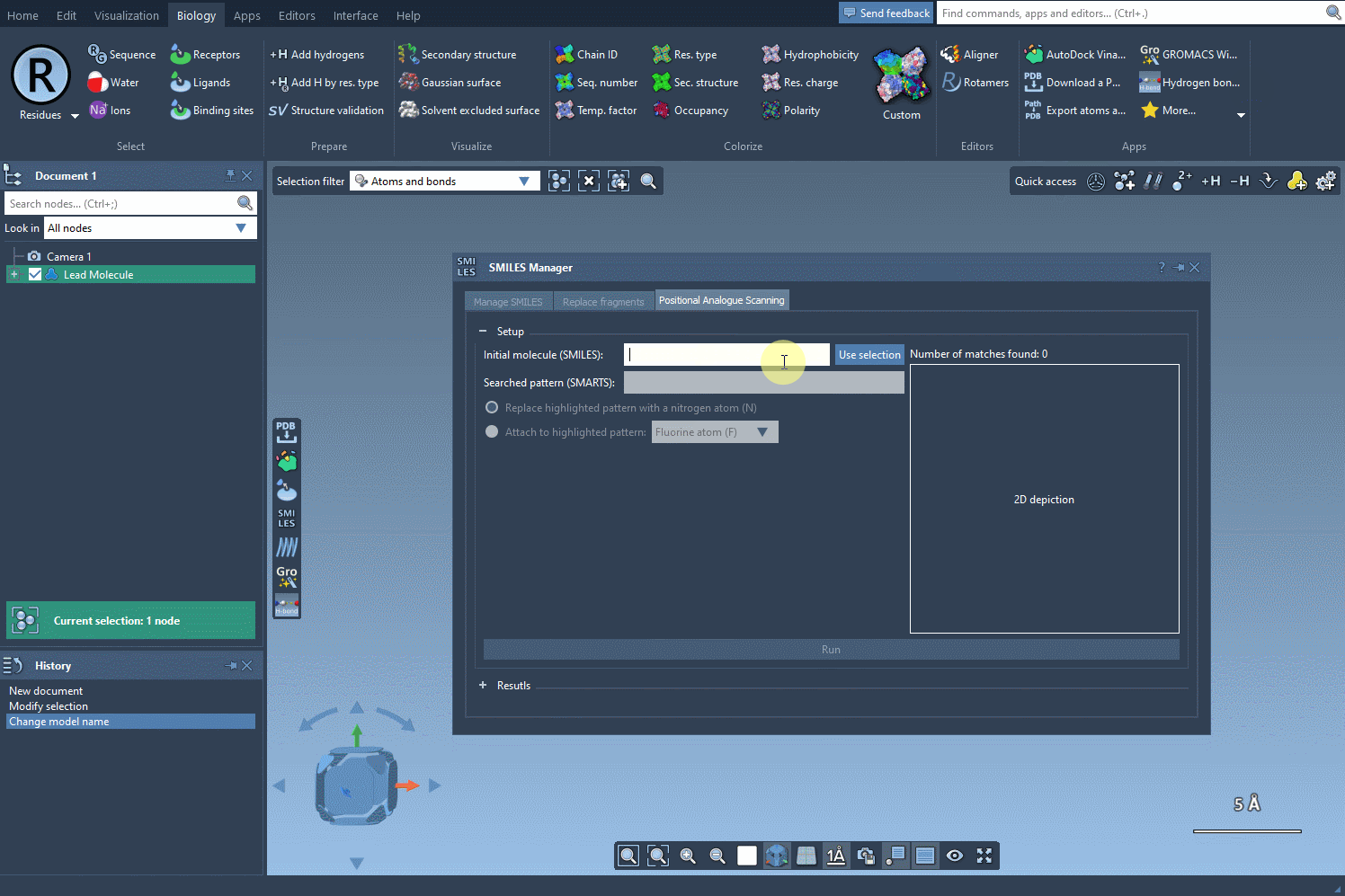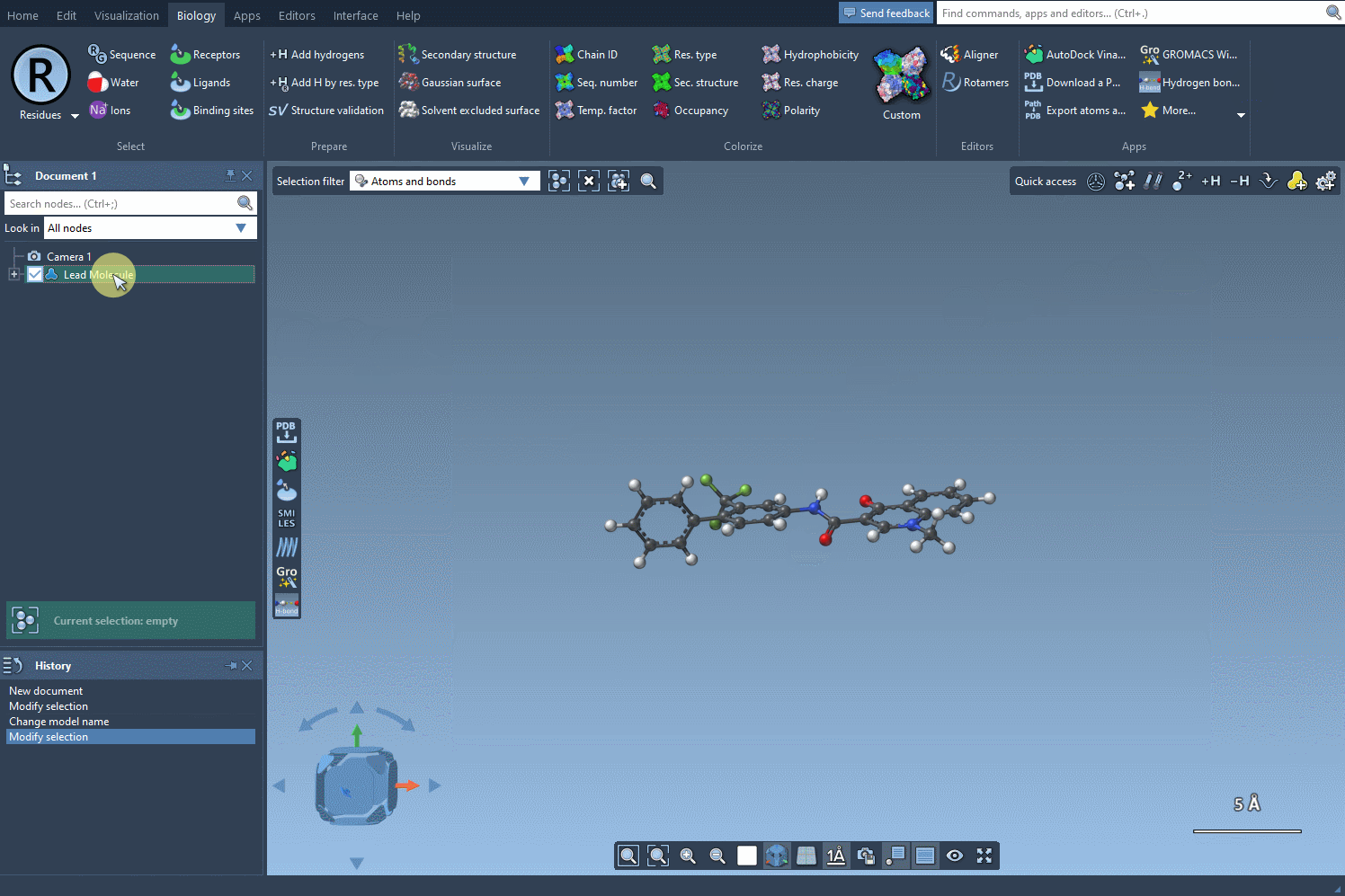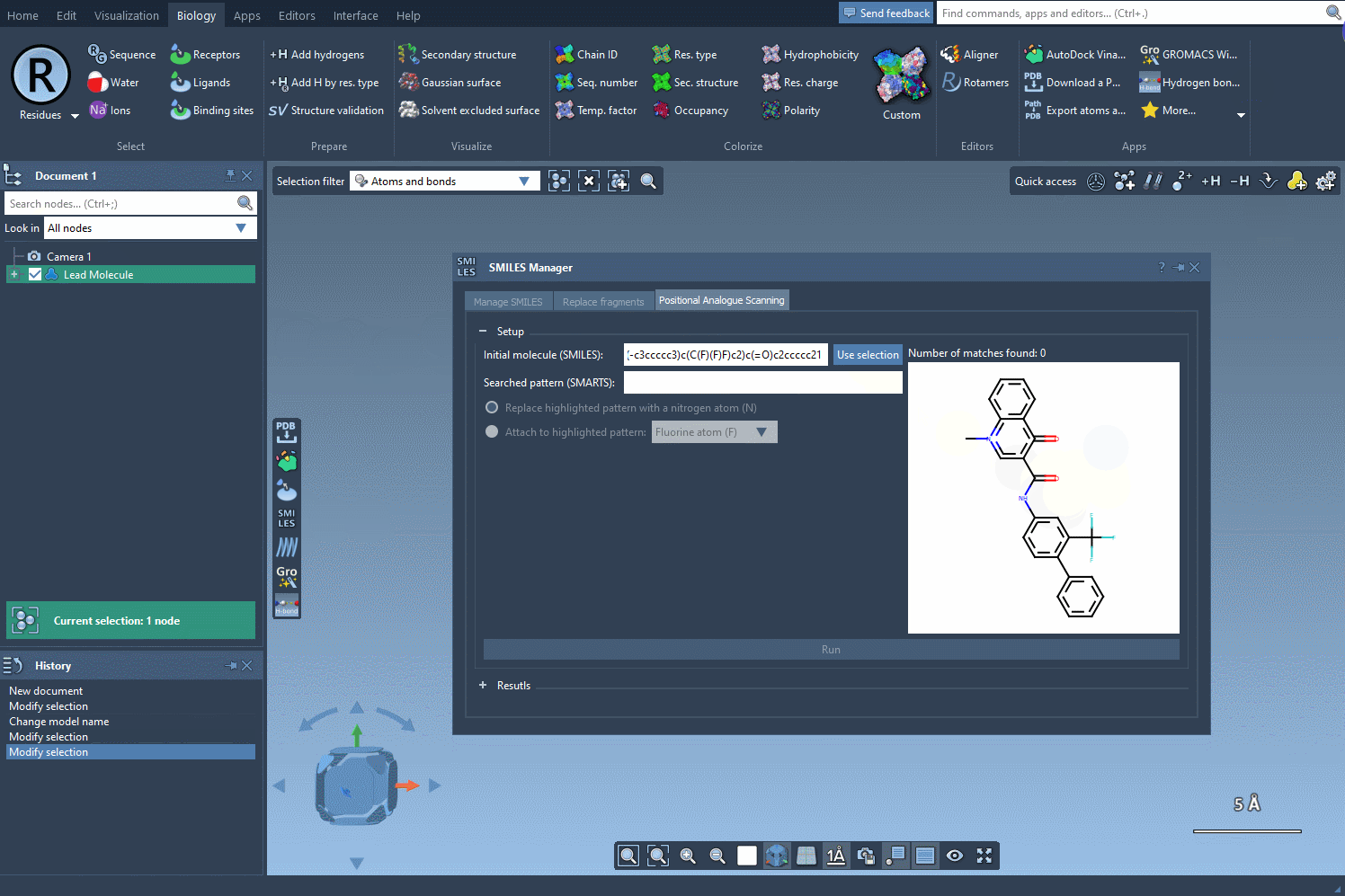
Why This Matters
This small feature makes a big difference when used regularly:
- Faster iteration: Go from visualization to analog generation in seconds.
- Fewer errors: Avoid copy-paste mistakes or outdated SMILES codes.
- Workflow integration: Seamlessly connect your design, analysis, and visualization steps without context-switching.
This approach works particularly well when using SAMSON for positional analogue scanning. Since this technique involves generating a series of analogs by modifying specific chemical positions, starting from an already visualized molecule lowers the barrier to exploration. You can spot patterns visually and decide on attachments or substitutions more intuitively.
Getting Started Is Easy
If you’re already using SAMSON, simply follow these quick steps:
- Select your molecule (or fragment) in the SAMSON document.
- Open SMILES Manager.
- Click Use selection.
Your molecule is now ready for analog generation, pattern search, docking prep, or any other downstream step in SAMSON.
It’s a small tweak, but it adds up — especially if you work with multiple compounds or need to prototype frequently. There’s no need to break your momentum just to re-format input data.
To explore more steps of the positional analog scanning process — such as identifying replaceable substructures, generating SMILES, or converting results to 3D — check out the full documentation here.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





