When analyzing molecular dynamics or preparing systems for enhanced sampling methods, it is often unnecessary—or even overwhelming—to export the entire atomic system. Molecular modelers frequently need to extract just specific atoms, like a ligand, or the residues lining a binding pocket, along a predefined unbinding pathway. This can be especially helpful for reaction coordinate generation, or when feeding meaningful input into free energy simulations.
The Export Along Paths extension in SAMSON makes this process easier than you might think. Here’s how to streamline your atom export process, focusing only on a chosen subset of atoms along computed paths.
Why export a subset?
Let’s say you’ve used Ligand Path Finder in SAMSON to determine the unbinding or binding trajectory of a ligand inside a protein pocket. You’ve optimized it using techniques like the parallel nudged elastic band method. The next logical step might be to perform free energy calculations. But for that, you may only need the ligand’s coordinates along the trajectory—exporting the entire complex for every frame could introduce unnecessary complexity.
Step-by-step guide to exporting a subset of atoms
Once your paths are ready and you’ve already loaded your sample system, here’s the key part that allows surgical precision:
- Open the Export Along Paths app via Home > Apps > All or search using Shift + E.
- Expand the Advanced panel in the app’s interface.
- In the Document view, select the atoms you’re interested in—this could be your ligand (like
TDG) or any set of residues. - Click Add. This defines a model to export. You can now see your selection listed with a name, which you can rename by double-clicking.
- Repeat the selection and definition process if you’d like to export multiple atom groups.
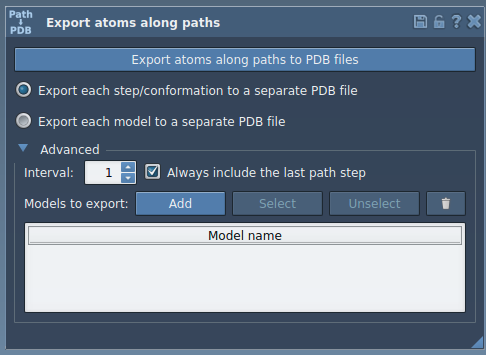
The application lets you rename the selection, click to highlight atoms in the Document view, or redefine the selection using the context menu or reset button.

Export settings
- Choose your preferred export format—either all frames in a single PDB file or each frame as a separate file.
- Select the corresponding path or paths from your document.
- Click Export atoms along paths to PDB files, and specify a destination folder and filename prefix.
This focused export allows you to work with only the atoms that matter for your downstream analyses—saving you time, storage, and potential preprocessing headaches.
Final thoughts
Exporting subsets of atoms along a defined molecular path may sound like a minor feature, but it can radically simplify tasks such as computing reaction coordinates, or feeding only necessary data into external simulation tools. If you’ve been exporting full molecular systems only to post-process them for these tasks, this feature can make your workflow far more efficient.
To dive deeper and follow the complete guide, including exporting all atoms and preparing the sample system, visit the full SAMSON documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. To get started, download SAMSON from https://www.samson-connect.net.





