Automatic symmetry detection in molecular structures can be a huge time-saver. But if you’re working with large biological assemblies, such as viral capsids or protein complexes with ambiguous layouts, automated tools might suggest several plausible symmetry groups. For structural biologists and molecular designers, that flexibility is great — until it adds uncertainty. Which axis is best? What if you already know the expected symmetry?
This is where manual symmetry group selection becomes essential. In this post, we’ll cover the pain behind leaving symmetry detection fully to algorithms and show how SAMSON’s Symmetry Detection Extension helps you regain control when automatically detected results are too broad or don’t match expectations.
Why Manual Selection?
In large complexes — for example, the protein structure 1B4B — SAMSON may detect multiple symmetry groups because different symmetrical arrangements can offer similar RMSD fitting results. However, you may:
- Already know the biological assembly is supposed to exhibit a particular symmetry (e.g.,
D3). - Need to prepare data for simulations that rely on a specific symmetry input.
- Want to speed up computations by extracting the correct asymmetric unit right away.
Luckily, the process to override or fine-tune automatic detection is simple and transparent.
How to Specify a Symmetry Manually
After launching the Symmetry Detection extension and loading your structure:
- Click Compute symmetry. SAMSON will detect and list possible groups.
- Instead of accepting the default group, scroll to the symmetry group interface.
- Pick the desired category (e.g., Dihedral) and desired order (e.g., 3 for D3) from the dropdown menus.
- Visual feedback instantly shows how the new selection maps onto your model.
This method gives you the precision to match your symmetry expectations to the actual detected geometry within the structure — minimizing artifacts and ensuring your decisions align with known biological information.
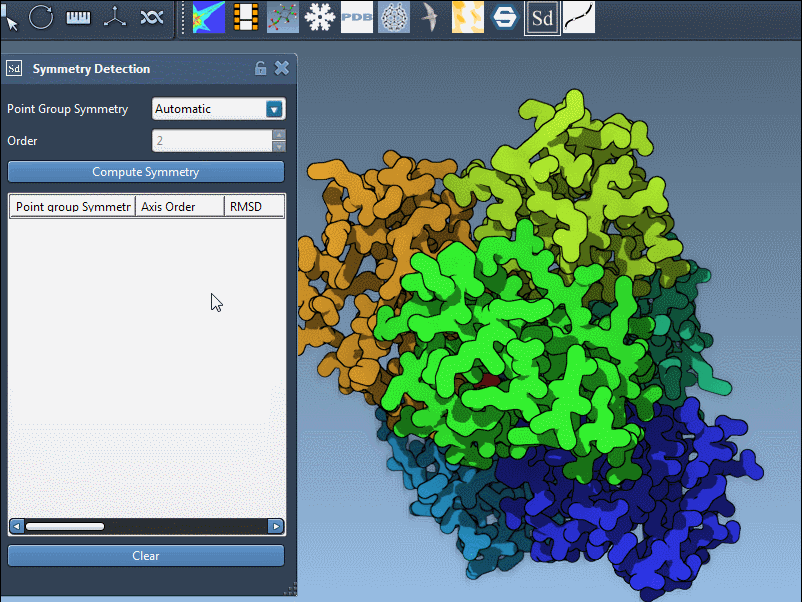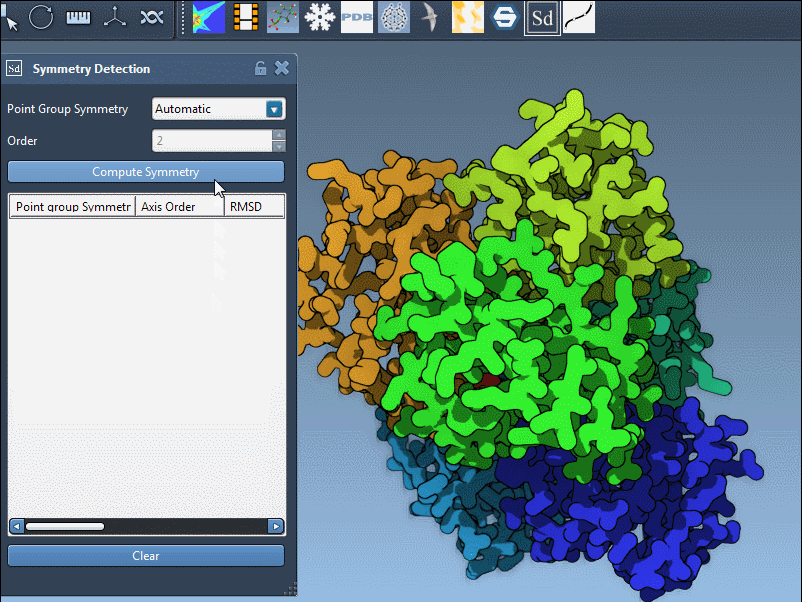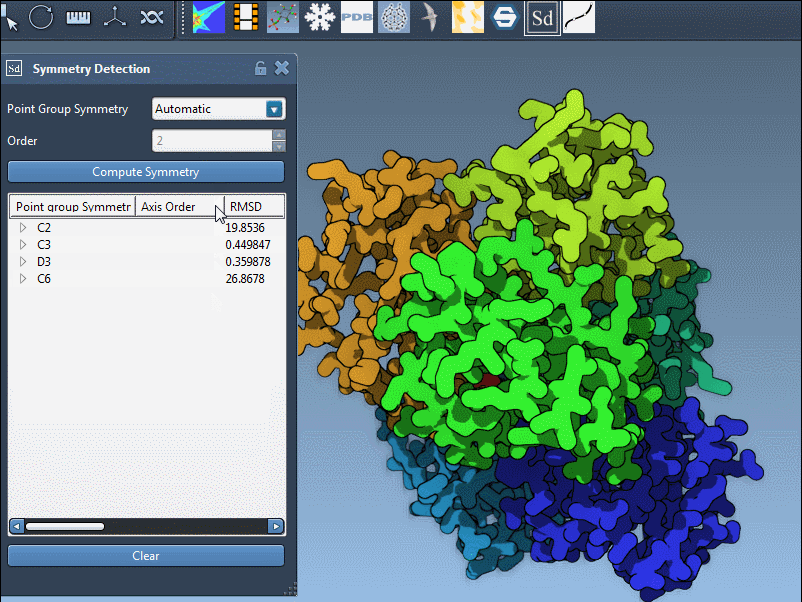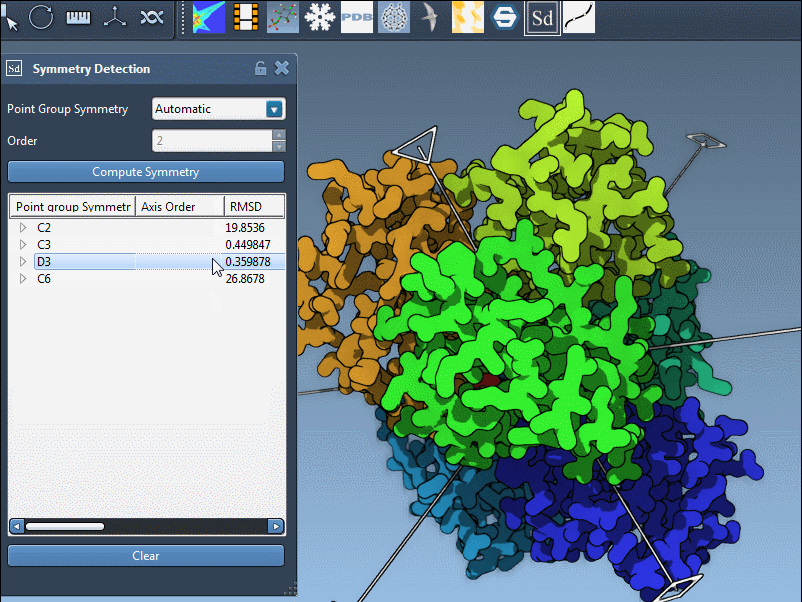
Bonus: Explore the Symmetry Axes
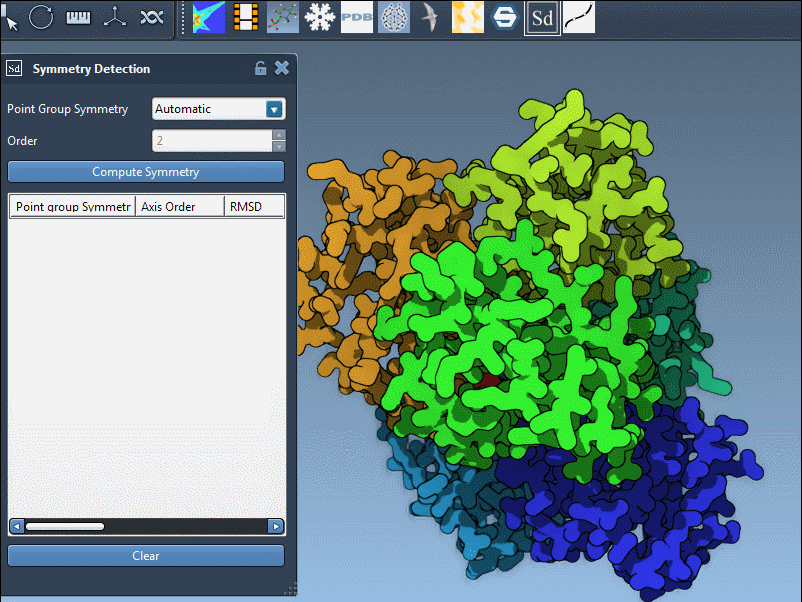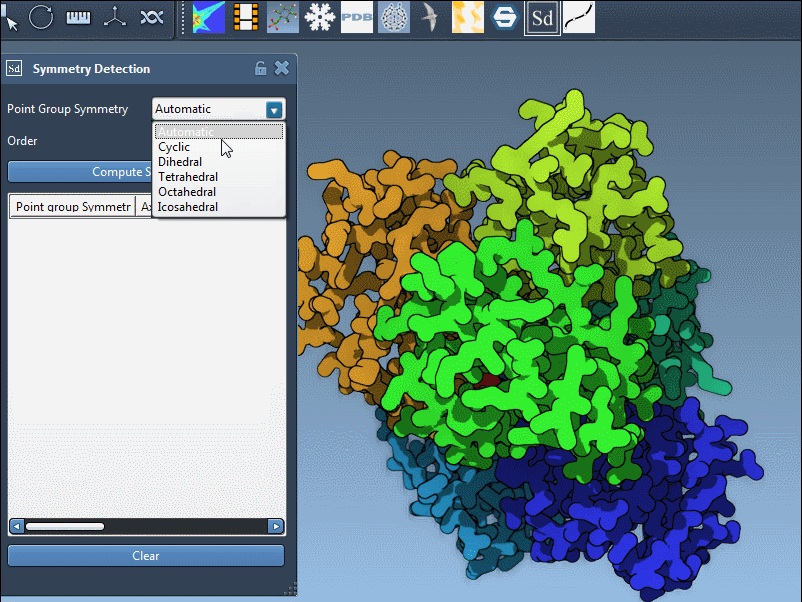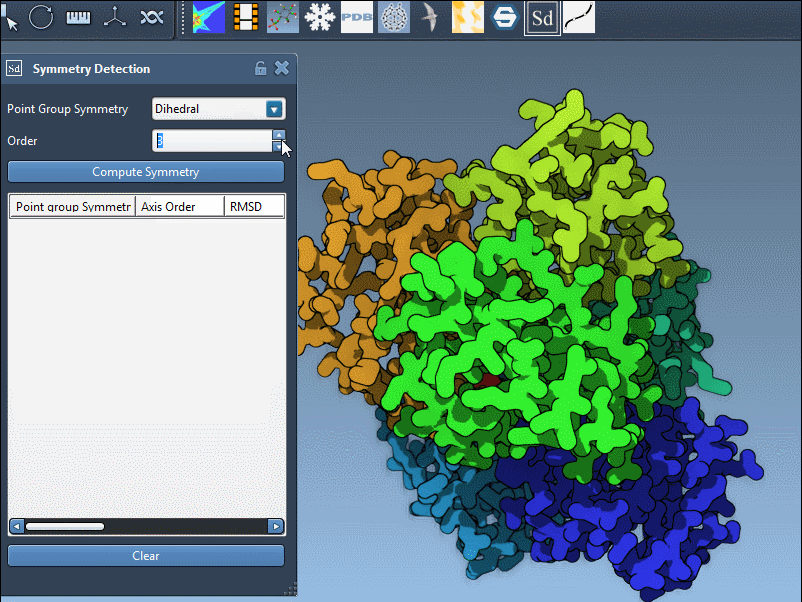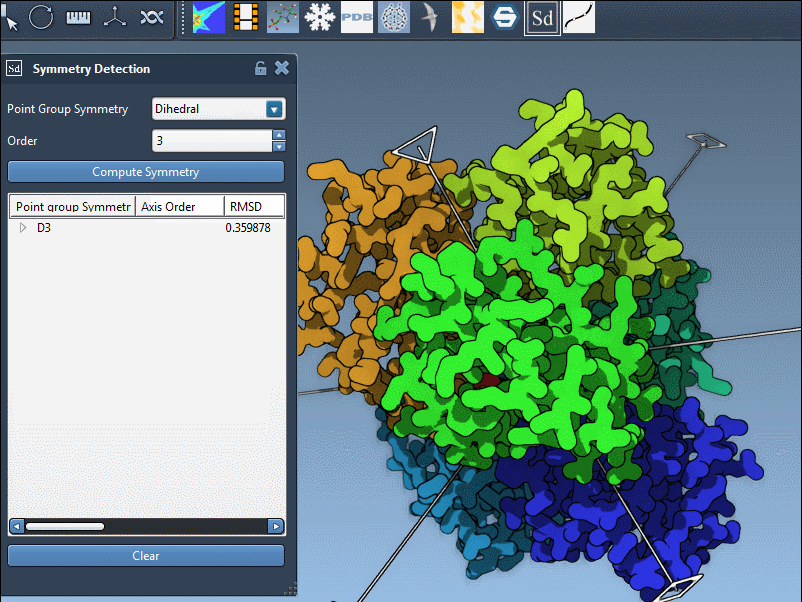
Once you choose a group, expand it to view individual axes. You’ll find tools like:
- Single-click: highlights that axis in bold
- Double-click: aligns your viewport along the axis
For the 1B4B structure, which has D3 symmetry, this can be really useful when preparing the system for symmetric docking, capsid modeling, or visualization exports.
Takeaways
If you work with large protein assemblies or viral structures, combining SAMSON’s automated tools with manual input is often the best strategy. It keeps your workflow fast — but with the ability to intervene when you know more than the algorithms do.
To learn more and start exploring symmetry detection in detail, visit the full documentation tutorial: https://documentation.samson-connect.net/tutorials/symmetry/computing-axes-of-symmetry-of-biological-assemblies/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





