Molecular modelers often face a recurrent frustration when studying large biomolecular structures: the lack of dynamic, visual insight into conformational transitions. This is particularly true for complex glycoproteins like the SARS-CoV-2 spike protein, which plays a central role in viral entry and infectivity.
While structural snapshots are helpful, understanding how such a protein transitions from a closed to an open state is critical for vaccine design, antibody development, and mechanistic studies. Fortunately, a detailed and visual representation of this motion is now accessible thanks to SAMSON, the platform for integrative molecular design.
Why the Spike’s Motion Matters
The spike (S) protein of SARS-CoV-2 undergoes a conformational change that enables the virus to bind to the ACE2 receptor on human cells. This motion — from a closed, inactive form to an open, binding-ready state — is a key target for preventative interventions like neutralizing antibodies.
Having a trajectory of this motion allows researchers to:
- Analyze intermediate states that might expose transient binding pockets
- Understand how neutralizing antibodies could interfere with the opening
- Design molecules that stabilize the closed (non-infectious) state
Downloadable Trajectories for Immediate Use
The SAMSON team provides several downloadable options for visualizing and analyzing the spike protein’s transition between the closed (PDB 6VXX) and open (PDB 6VYB) states:
- PDB trajectory files
- Single PDB file
- SAMSON format file (includes animation and structure)
The animations help visualize the entire transition path:
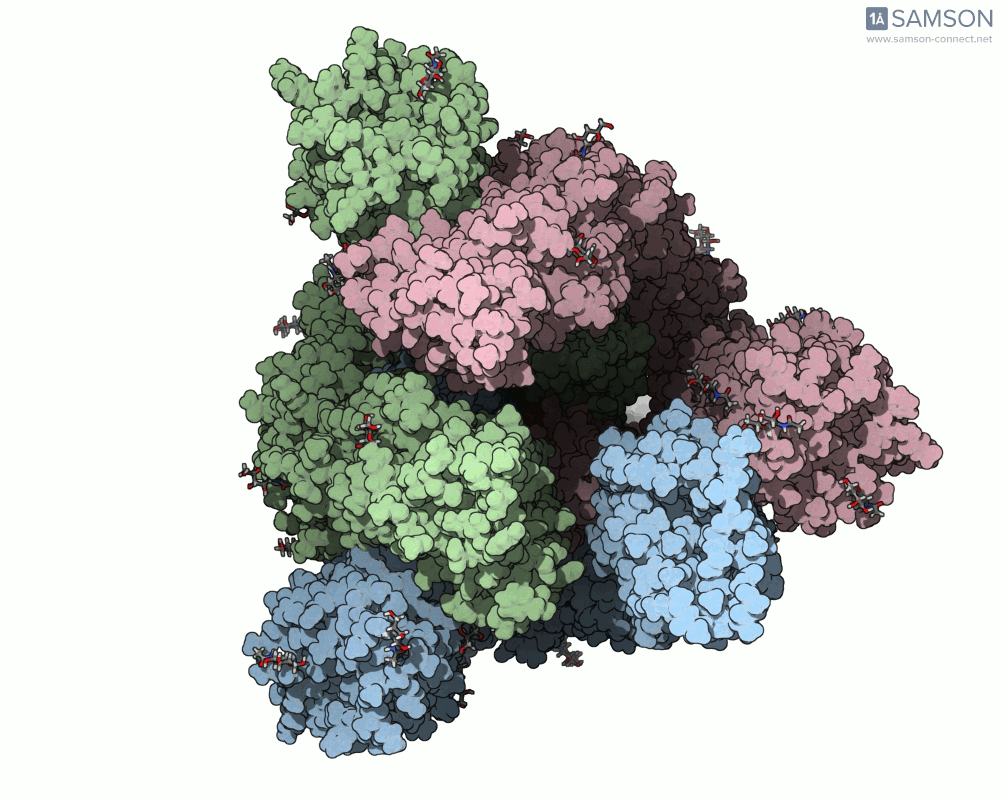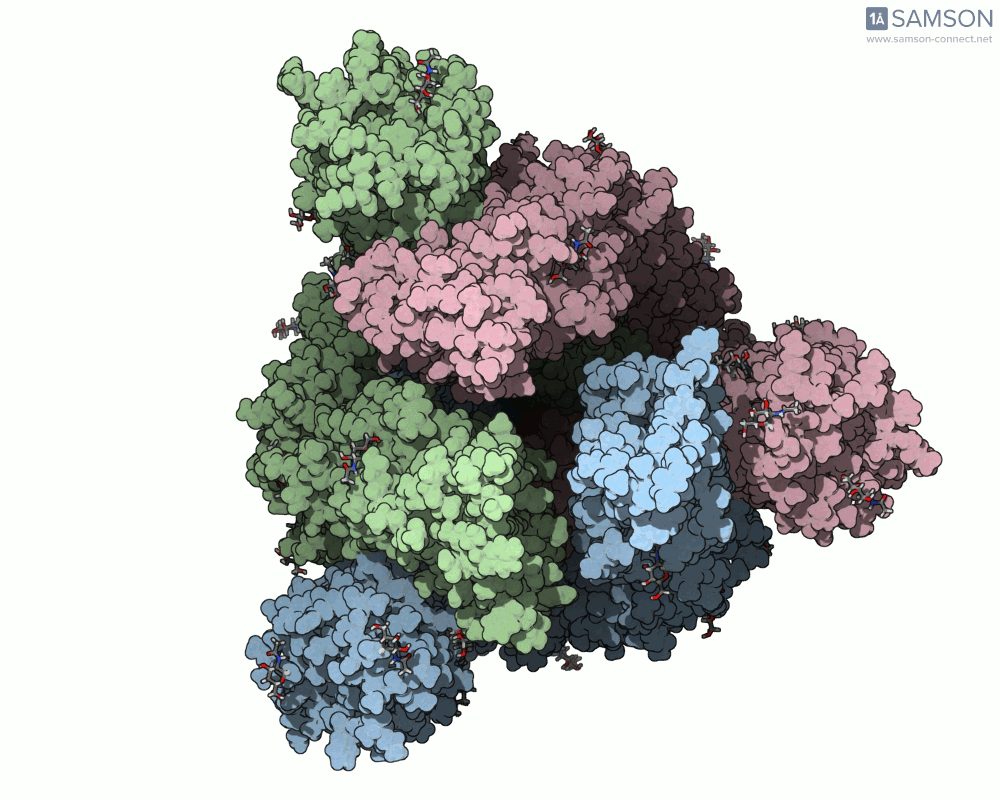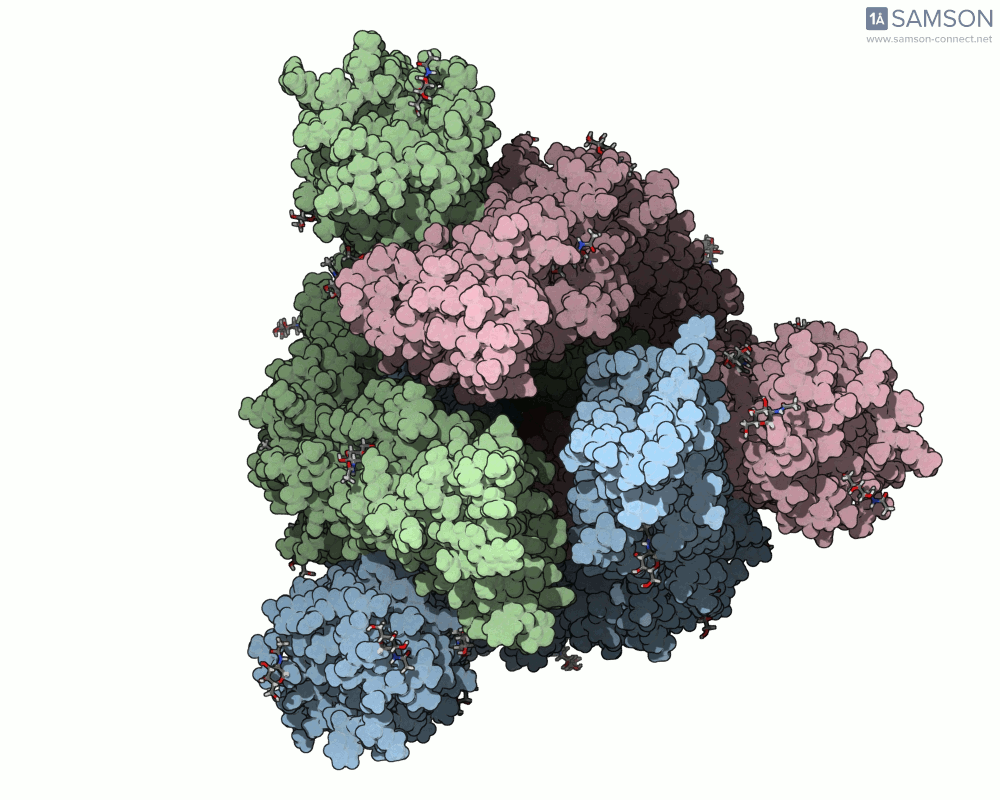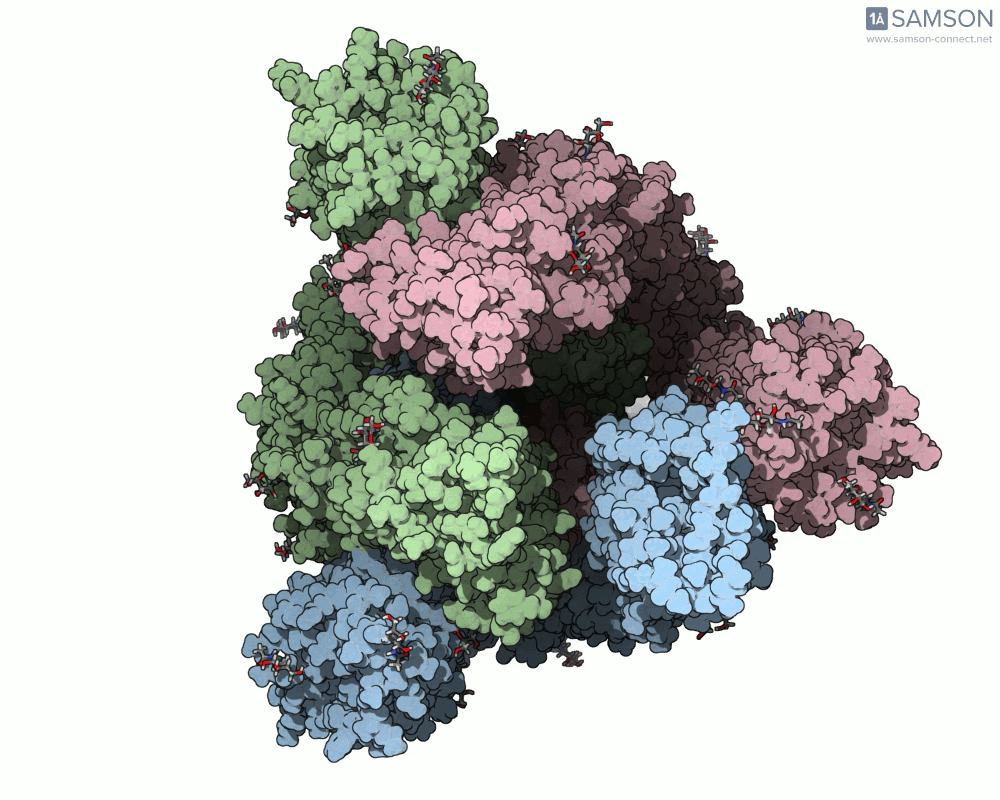
How the Animation Was Computed
The motion is generated using a two-step pipeline in SAMSON:
- ARAP Interpolation: Provides an initial trajectory by interpolating between the open and closed states.
- P-NEB (Parallel Nudged Elastic Band): Refines the pathway to a more physically plausible transition.
Both modules are available for free on the SAMSON Connect platform. Trajectories are generated using the structure of the open conformation as a reference and follow through intermediate minimizations to manage differences in residue counts between states.
What You Can Do With It
Whether you are simulating antibody interactions or studying glycan shielding, these trajectories can be used as input for further computational experiments. For example, you could select individual frames of the trajectory for docking studies or compute energetics along the motion pathway using additional SAMSON tools or your own force field implementations.
Even though the path has not been experimentally validated, it can offer a meaningful starting point — especially when experimental intermediate structures are lacking.
To Learn More
See the full documentation, including animated figures, downloadable files, and technical details, at the original article: Visualizing the Opening Motion of the SARS-CoV-2 Spike.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON and get started at https://www.samson-connect.net.





