In molecular modeling, building a lipid bilayer around a membrane protein is often a necessary step for meaningful simulations. Whether you’re preparing a system for molecular dynamics or just visually analyzing structural interplay, constructing a realistic lipid environment can be time-consuming. This process often involves scripting, trial-and-error, or external tools that can break your workflow.
Thankfully, with the Molecular Box Builder extension in SAMSON, you can build a lipid bilayer around your protein directly within the same platform. No scripts. No format conversions. Just a guided user interface that walks you through each step.
Aligning the Protein Structure
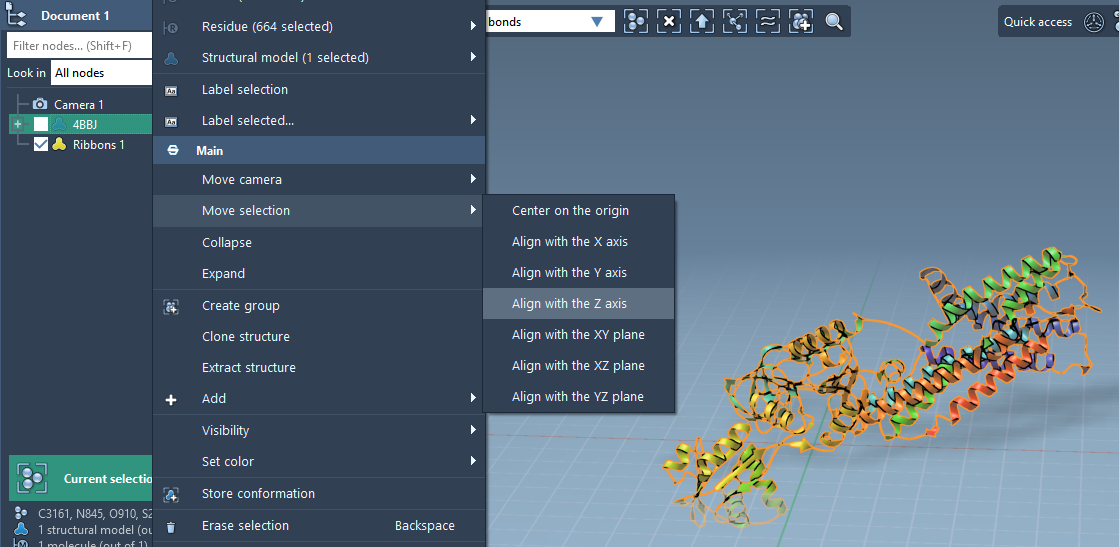
The first step is to reorient your membrane protein so that it sits perpendicular to the bilayer plane. This is done by aligning the protein to the Z-axis and centering it at the origin.
- Right-click the protein in the Document view.
- Select Move selection > Align with Z axis.
- Then choose Move selection > Center on the origin.
Insert the Lipid Molecule
Next, you select a lipid molecule to populate your bilayer:
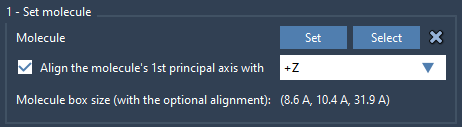
- Import or load the lipid model you want to use, such as POPC.
- Select it in the Viewport or Document view, then click Set in the Molecular Box Builder app.
- Choose to align it with the
+Zaxis.
Define the Region to Fill
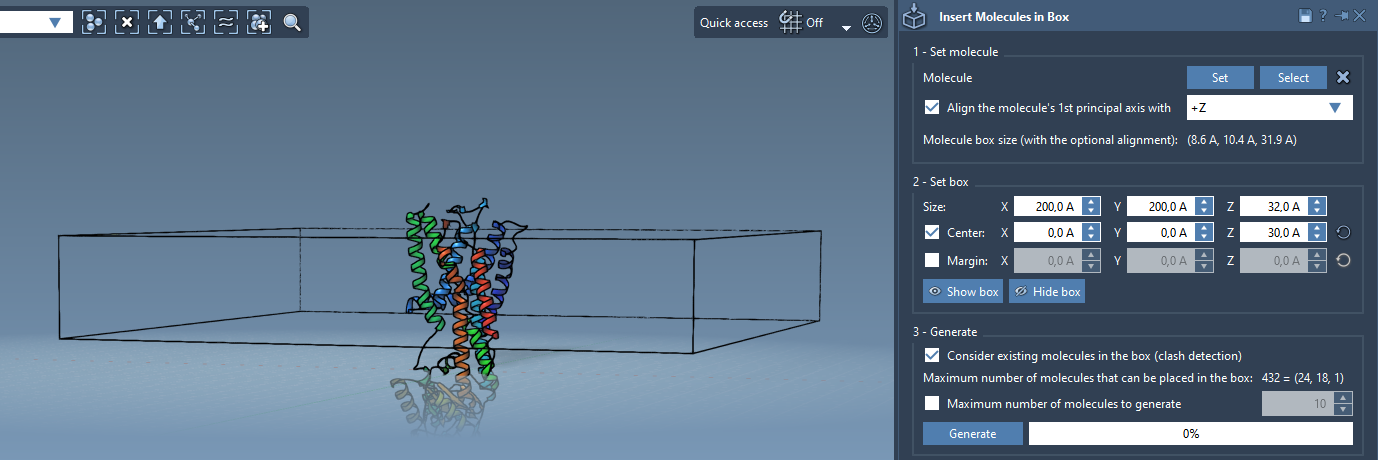
Create the box where lipid molecules will be inserted:
- Center the box around the protein structure by enabling the Center option.
- Specify box dimensions in
X,Y, andZto include the protein and a mono- or bilayer of lipids. - Optionally, define margins to prevent overlaps or adjust lipid density.
Building the Bilayer
To insert lipids without overwriting the protein atoms:
- Enable the Consider existing molecules in the box option.
- Click Generate to create the first lipid layer.
- Shift the box slightly along the
Z-axis to insert the second layer. - Set the lipid alignment to
-Zfor the second layer and generate again.

Next Steps
That’s it — you now have a lipid bilayer assembled around your membrane protein, ready for minimization or simulation. You can continue by solvating the system or equilibrating it with tools like the GROMACS Wizard, which is also available in SAMSON.
To learn more, visit the full Molecular Box Builder documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. Get SAMSON at www.samson-connect.net.





