Running molecular simulations at the scale of full biological assemblies can be time-consuming and computationally intensive. But often, nature provides a shortcut: symmetry. If your system has symmetry, you might not need to simulate the entire structure. Instead, you can focus on the repeating asymmetric unit — vastly reducing the system size without losing meaningful information.
The Symmetry Detection extension in SAMSON offers an efficient workflow to identify and visualize symmetries in protein complexes and large molecular assemblies. Beyond visualization, this tool helps you streamline your simulation setup. Here’s how molecular modelers can benefit from it.
Why simulation size matters
Let’s say you are preparing a molecular dynamics run to study a viral capsid. Simulating the full complex requires substantial computational resources, which may be infeasible for long time scales. But if the capsid has icosahedral symmetry, you might be able to extract one asymmetric unit and simulate that alone — provided symmetry is properly identified and respected by the force field and boundary conditions.
Using SAMSON to find the asymmetric unit
Once you load your structure in SAMSON (via Home > Fetch or from a local file), follow this procedure:
- Run the Symmetry Detection extension (Home > Apps > Biology > Symmetry Detection).
- Click Compute symmetry.
- The detected symmetry axes are rendered in the viewport for review.
In the case of icosahedral systems like 3NQ4, the app reveals all 2-fold, 3-fold, and 5-fold symmetry axes:
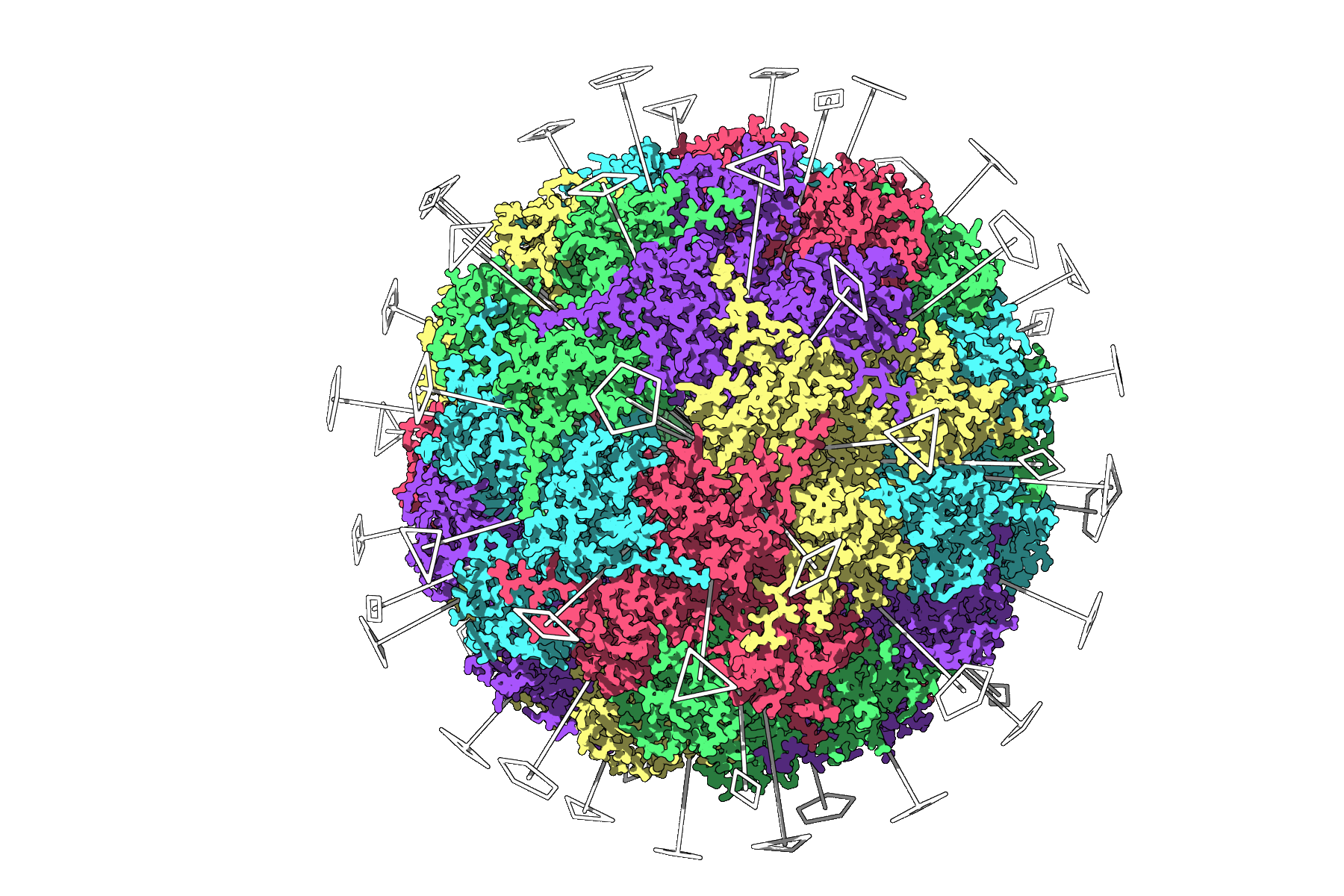
From here, you can choose the symmetry group that best fits your setup. For systems with multiple possible symmetries, like 1CHP, you can compare options and see which yields the lowest root-mean-square deviation (RMSD) — a good proxy for symmetry quality.
Evaluating and selecting symmetry groups
SAMSON lets you explore detected groups quickly. Click a group to highlight its primary axis. You can also manually specify a group if you know the expected symmetry, such as D3 for 1B4B:
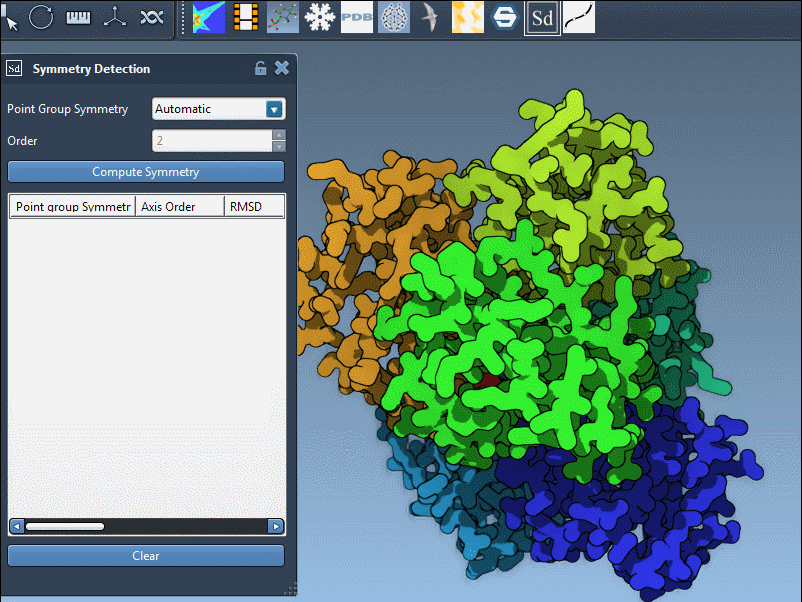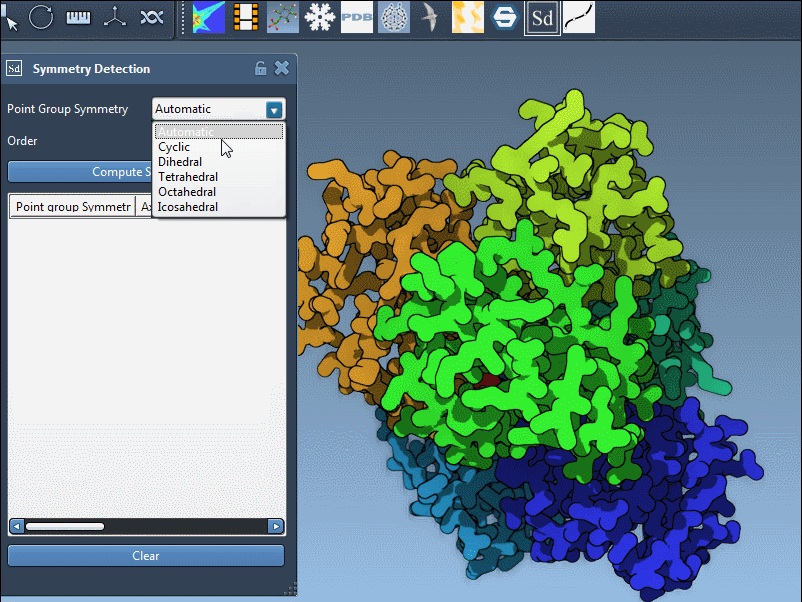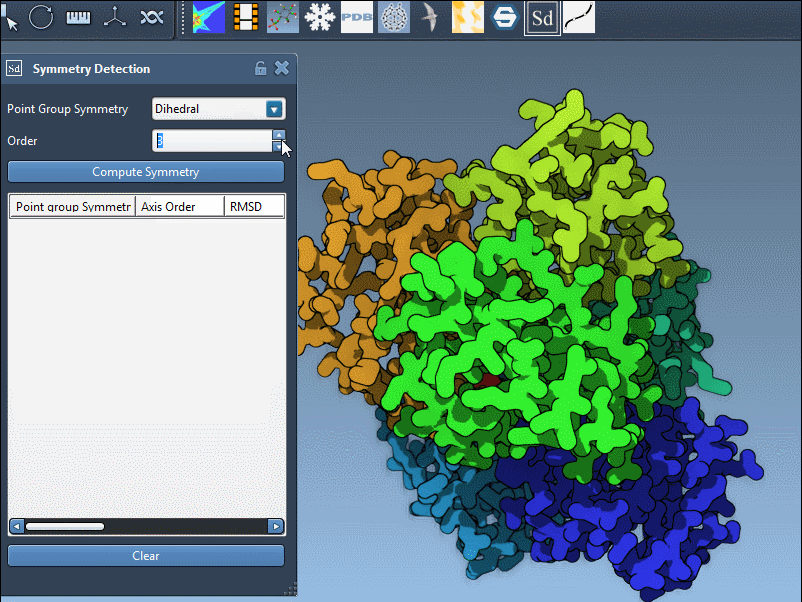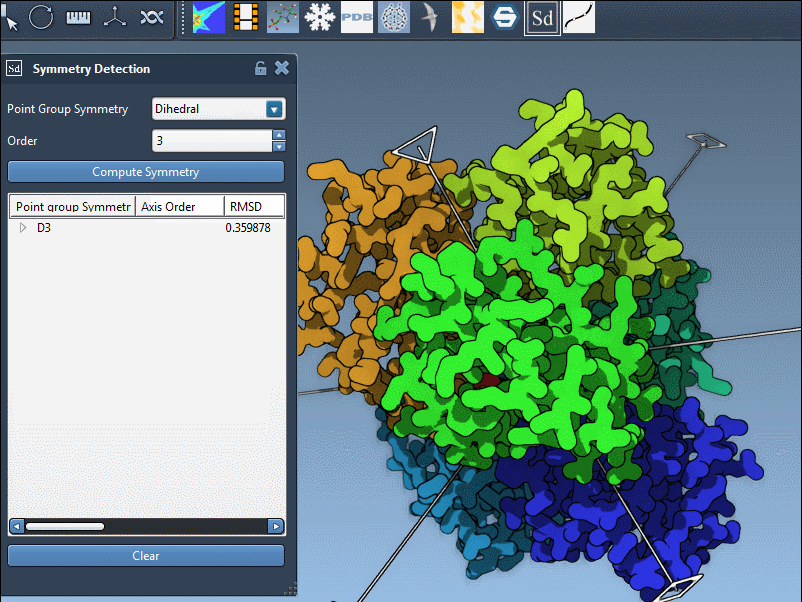
Clicking on an axis shows it in bold, while double-clicking aligns the camera along the symmetry axis. This visual feedback is useful when preparing publication figures or confirming structural repeat units before extraction.
Exporting the asymmetric unit
After selecting the symmetry group and axis of interest, you can isolate and export the asymmetric unit for simulation. This unit captures the unique molecular environment without redundant symmetric copies. It’s a significant time and resource saver, particularly in large systems with cubic or dihedral symmetry.
This approach is applicable in many contexts:
- Protein complex modeling
- Design of symmetric ligands for multivalent binding
- Nanoparticle design based on protein building blocks
Getting the most out of visualization
For clarity in presentations or publications, combine symmetry axes with Ribbon or Surface representations, and consider coloring separate chains or asymmetric units differently. SAMSON also allows you to capture viewport snapshots directly.
Conclusion
Using symmetry detection in SAMSON isn’t just about visualization — it’s a practical shortcut to more efficient simulations. If your molecular system contains symmetries, finding and exploiting them can give you faster results and cleaner models.
To learn more and explore additional features, visit the official documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





