Working with molecular models often involves tweaking structures to better fit expected configurations, eliminate artifacts, or explore structural variations. But what happens when you’re already mid-simulation, and you realize a modification is needed—say, removing a misplaced hydrogen, or adding a missing functional group?
Traditionally, this might mean stopping the simulation, editing the model in a pre-processing stage, and rerunning everything. In SAMSON, however, this is no longer the case when using the Universal Force Field (UFF): you can delete and add atoms on the fly, and SAMSON will automatically re-perceive the molecule accordingly. This means the force field adapts—live—as you edit. 🧬
Why is this important?
Being able to modify molecular structures without restarting simulations saves time and helps explore structural hypotheses quickly. It also allows newcomers and experts alike to use live molecular editing as a tool for learning, prototyping, or debugging force field interactions in real-time.
How to Delete and Add Atoms during a UFF Simulation
Once your simulation with UFF is running in SAMSON, here’s how to remove or insert atoms without breaking stride:
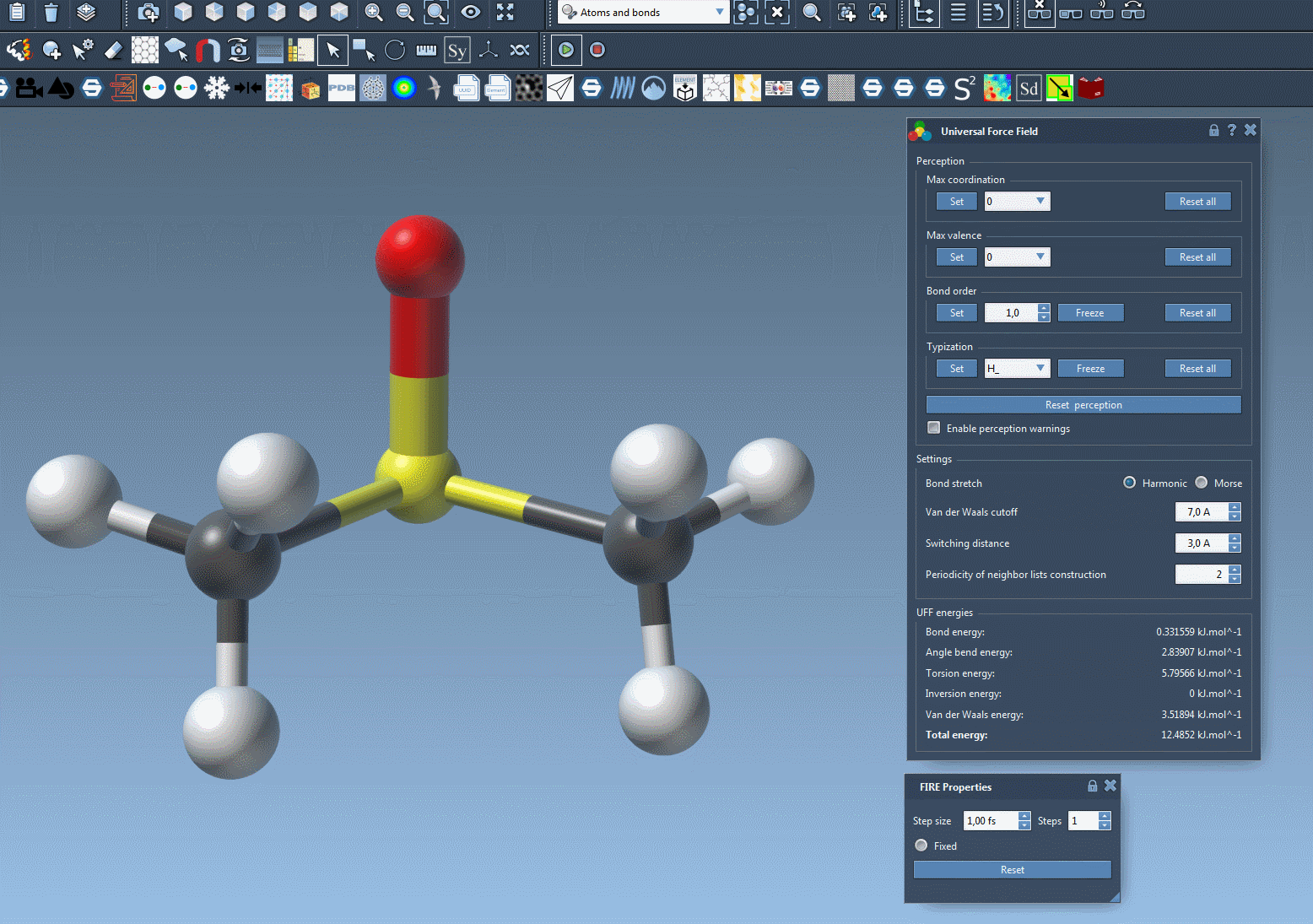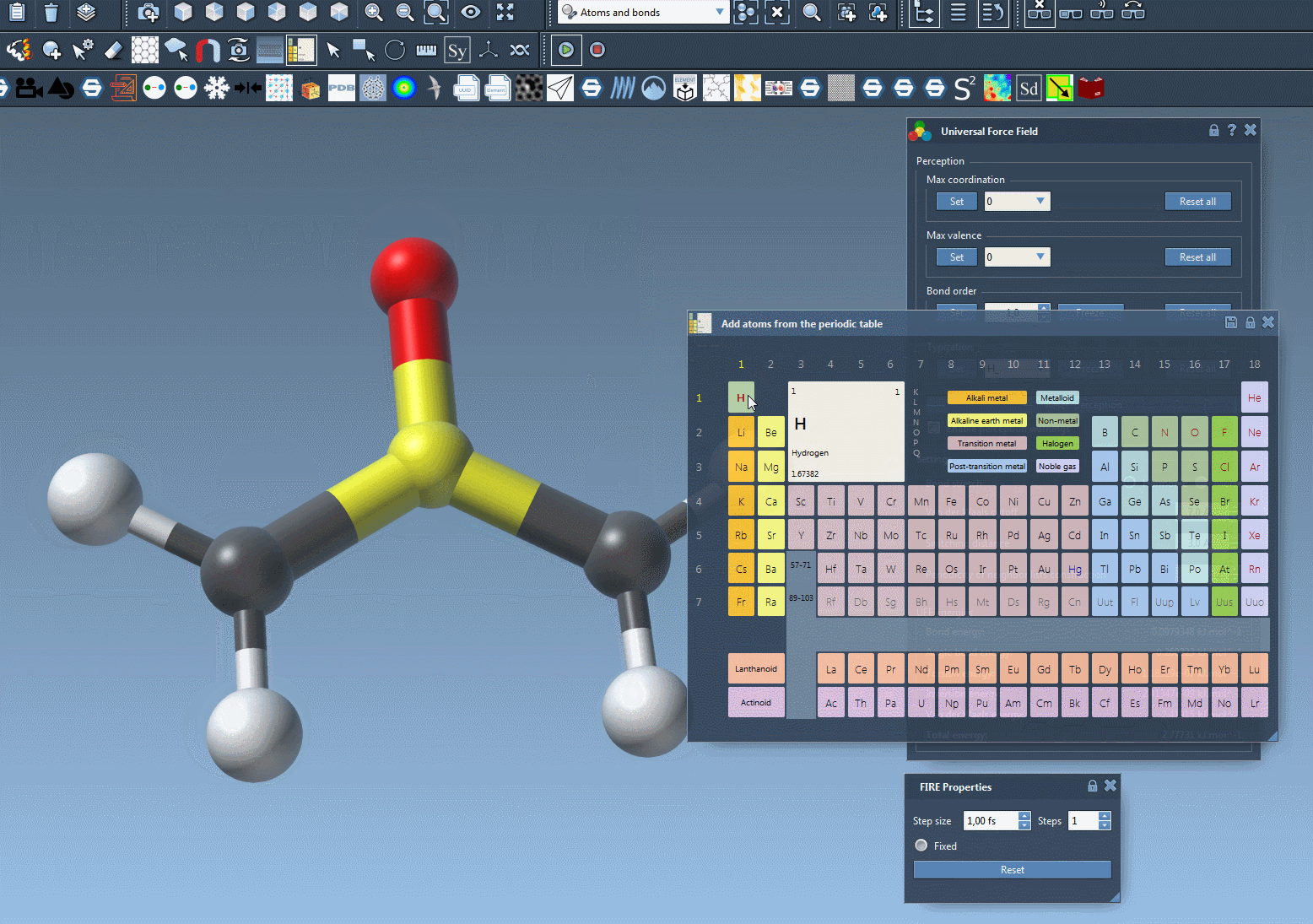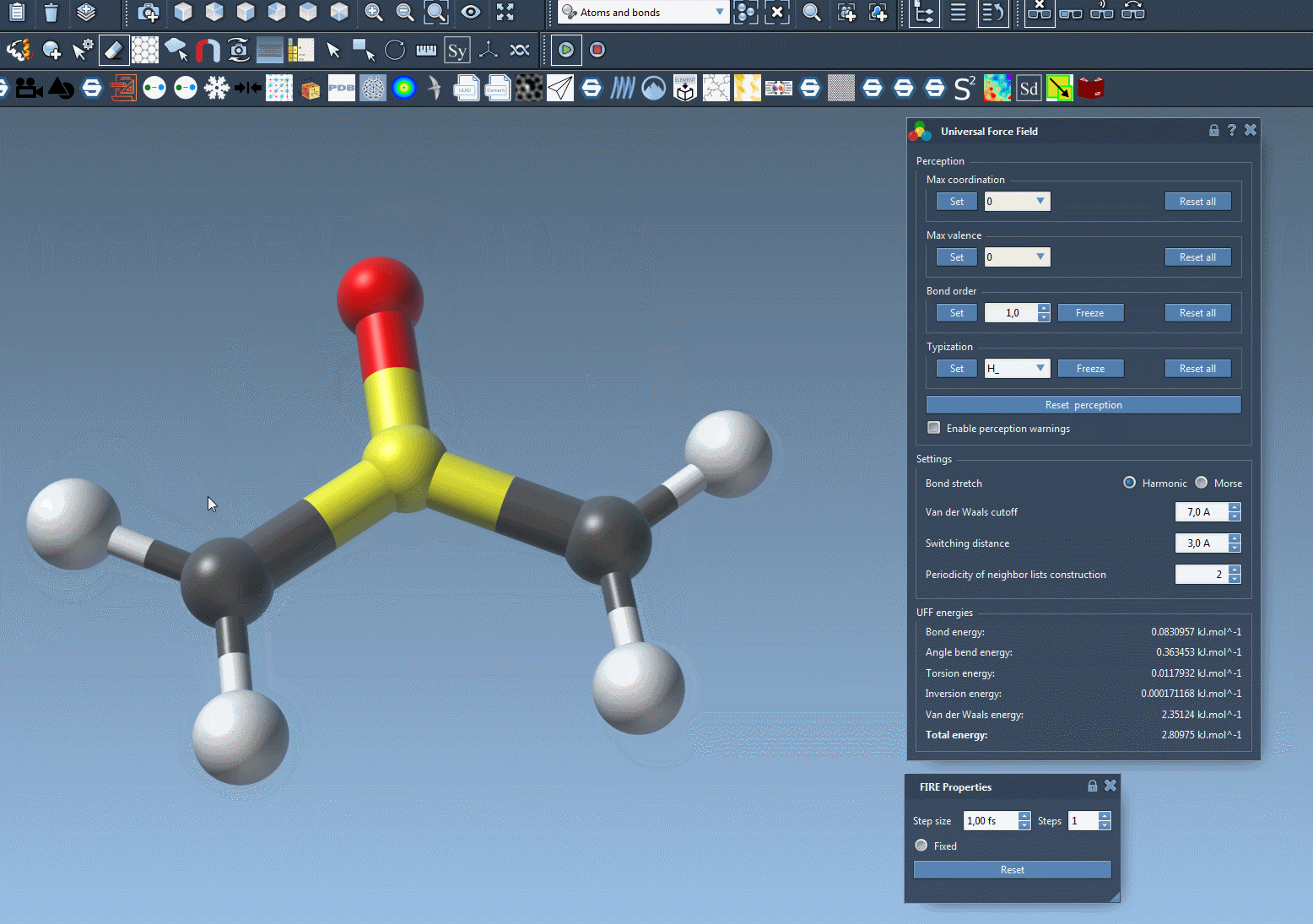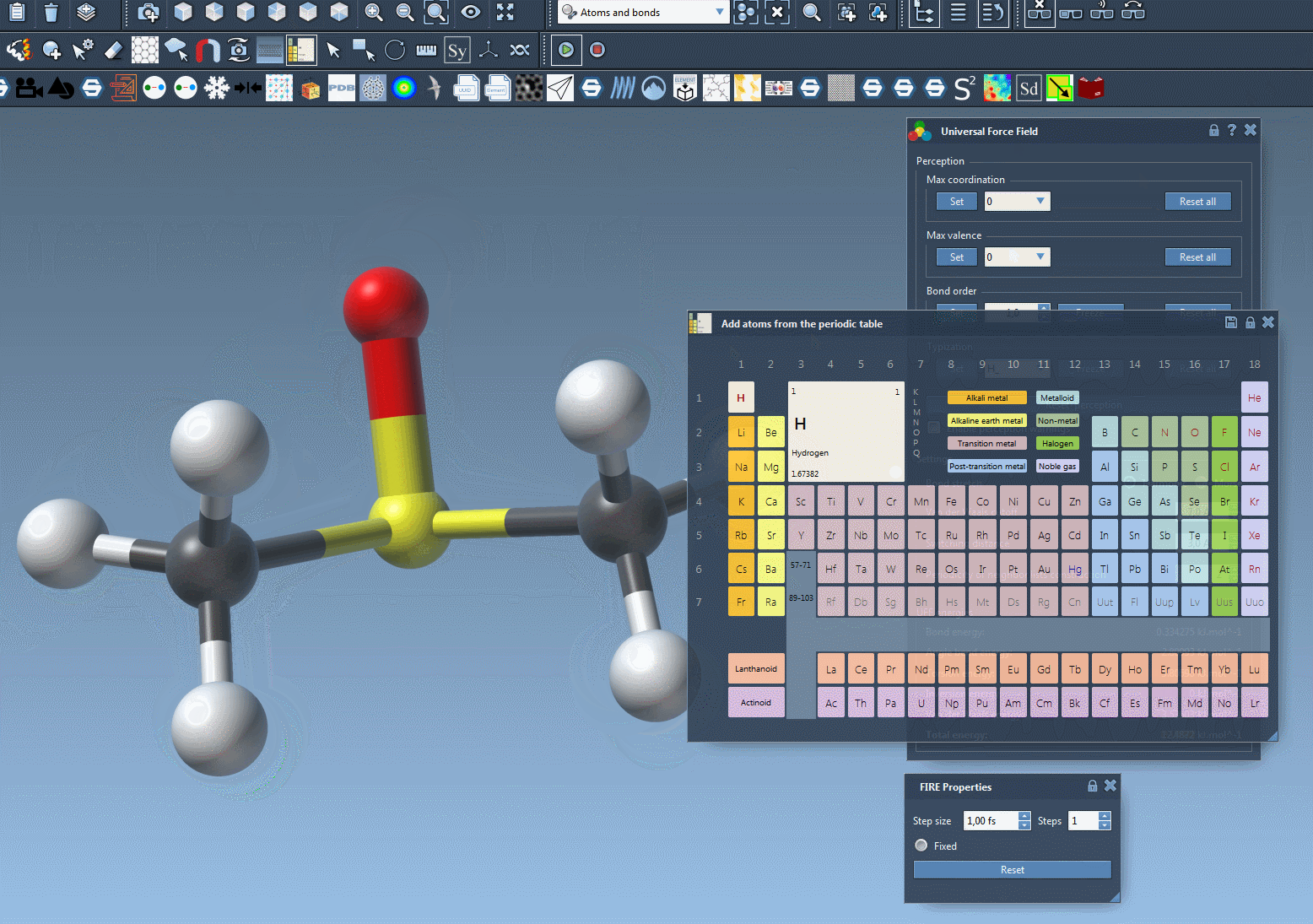
📎 Deleting Atoms
- Select the eraser editor (visible as a rubber icon in SAMSON’s editor toolbar).
- Click on the atom(s) you wish to delete.
SAMSON will automatically update the internal representation of the molecule and re-calculate atom types, bond orders, and other parameters based on the changed topology.
➕ Adding Atoms
- In the periodic table editor, choose the element you want to add (e.g. Carbon, Oxygen, etc.).
- Click on an existing atom in your model to which the new atom will be bonded.
- While holding down the mouse, move the cursor to define the bond length and orientation. SAMSON will preview the new atom in transparency.
- Release the mouse button to confirm the addition.
The new atom becomes immediately part of the structured molecular system. Typization (atom type, coordination, valence) and interactions are calculated instantly by the UFF module.
Automatic Typization and Consistency Checks
The great part is that SAMSON doesn’t just insert or remove atoms—it also runs automatic structure perception to update bonding and energetics. If inconsistencies are detected—such as valence violations or unassigned atom types—a warning message may appear. This helps you quickly identify if further adjustments are needed for the model to remain valid.
Use Cases and Practical Tips
- Fixing structures: Gaps or misassigned atoms from imported files can be manually corrected.
- Organic synthesis prototyping: Try modifying side chains and observe real-time force field responses.
- Demonstrations and teaching: Show how molecular mechanics responds to topological changes live in class or workshops.
Things to Keep in Mind
- Only atoms bonded to existing atoms are considered part of the simulation—random floating atoms aren’t connected automatically.
- You can add additional complexity by customizing typization, but for most users, the automatic perception is sufficient.
To explore more on how to build molecules in SAMSON, including interactive tutorials, check out the User Guide: Building molecules.
Live editing in UFF simulations is a feature that blends modeling flexibility and simulation rigor—helping you stay focused and creative without interrupting your workflow.
To learn more about running UFF simulations and customizing parameters, visit the UFF tutorial documentation page.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





