If you work with protein-ligand systems, a common challenge is capturing the movement of a ligand along a path — for example, its unbinding from the active site — and exporting this data in a format usable for downstream tasks like free energy profiling 🧪 or molecular visualization 🎥.
SAMSON’s Export Along Paths extension offers a straightforward solution to this. It enables exporting atomic coordinates along predefined paths, which is especially useful after computing ligand trajectories using tools like Ligand Path Finder or optimizing them with methods like the parallel nudged elastic band.
In this guide: Export only the ligand along its path
Rather than exporting all atoms in a large macromolecule, you might just want to focus on the ligand and track its atomic positions as it moves. Here’s how to do that efficiently with SAMSON.
1. Load a Sample System
To try this out, SAMSON provides a sample document containing the protein Lactose permease (PDB: 1PV7) with its ligand Thiodigalactosid (TDG), and two unbinding paths.

2. Launch the Export Along Paths App
In SAMSON, go to Home > Apps > All > Export Along Paths. You can also quickly find it using Shift + E.
3. Select a Subset: Just the Ligand
To focus only on the ligand:
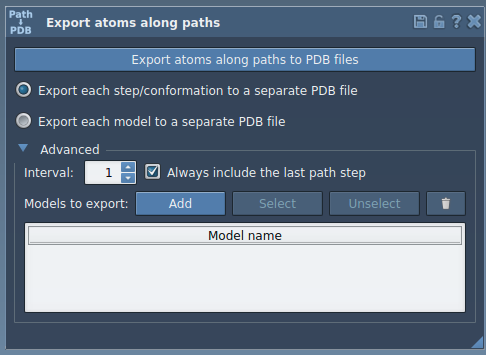
- Expand the Advanced panel in the Export Along Paths app.
- In the Document View, select the ligand (e.g.,
TDGmolecule). - Click Add to define the ligand as a model for export.

Named models appear in a table inside the app. You can rename them, select/unselect atoms, reset the selection, or add more molecules.

4. Export the Ligand’s Trajectory
- Choose whether to export as one PDB file (all frames) or as separate PDB files (each frame = one file).
- Select one or more paths in the Document View.
- Click Export atoms along paths to PDB files.
The app will prompt you to choose a folder and filename prefix.
Why this helps
Exporting only the ligand’s coordinates along a path is invaluable for molecular simulation tasks:
- Create reaction coordinates for free energy calculations.
- Track ligand motion during unbinding or binding events.
- Generate intermediate conformations for enhanced sampling.
- Visualize flexible ligand behavior without the background of the entire protein.
To explore other features — such as exporting all atoms, tweaking export intervals, or setting up multiple models — visit the full documentation.
👉 Learn more in the official documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON from https://www.samson-connect.net.





