A common challenge in molecular modeling is evaluating how small structural changes in a molecule impact properties like binding affinity or interaction patterns. This is particularly relevant in drug design, where exploring analogs can reveal promising candidates while eliminating ineffective ones.
Positional analogue scanning is one technique that simplifies this process. Rather than manually editing molecules to try out different substructures, you can automatically generate analogs by replacing specific atomic patterns — saving time and effort. In this post, we show how to do exactly that within the SMILES Manager extension of SAMSON, using a simple example that demonstrates the practical benefits of this approach.
Why replace substructures?
Suppose you’re working with a compound and you suspect that changing an aromatic carbon might affect its interaction with a target protein. Traditionally, you might manually draw variations of the molecule or run scripts outside your modeling environment — both time-consuming and disconnected from a seamless workflow.
Instead, SMILES Manager lets you define that structural motif (e.g., an aromatic carbon), replace it with atoms like nitrogen or attach groups such as fluorine or methyl, and instantly generate new analogs — all inside your modeling environment. It’s both fast and flexible.
How it works in SAMSON
After defining your initial molecule by entering its SMILES code or selecting it directly in the SAMSON document, navigate to the substructure pattern input field. For example, to target aromatic hydrogens, use the SMARTS pattern [cH]. SAMSON will automatically highlight every matching position in your molecule, guiding you visually.

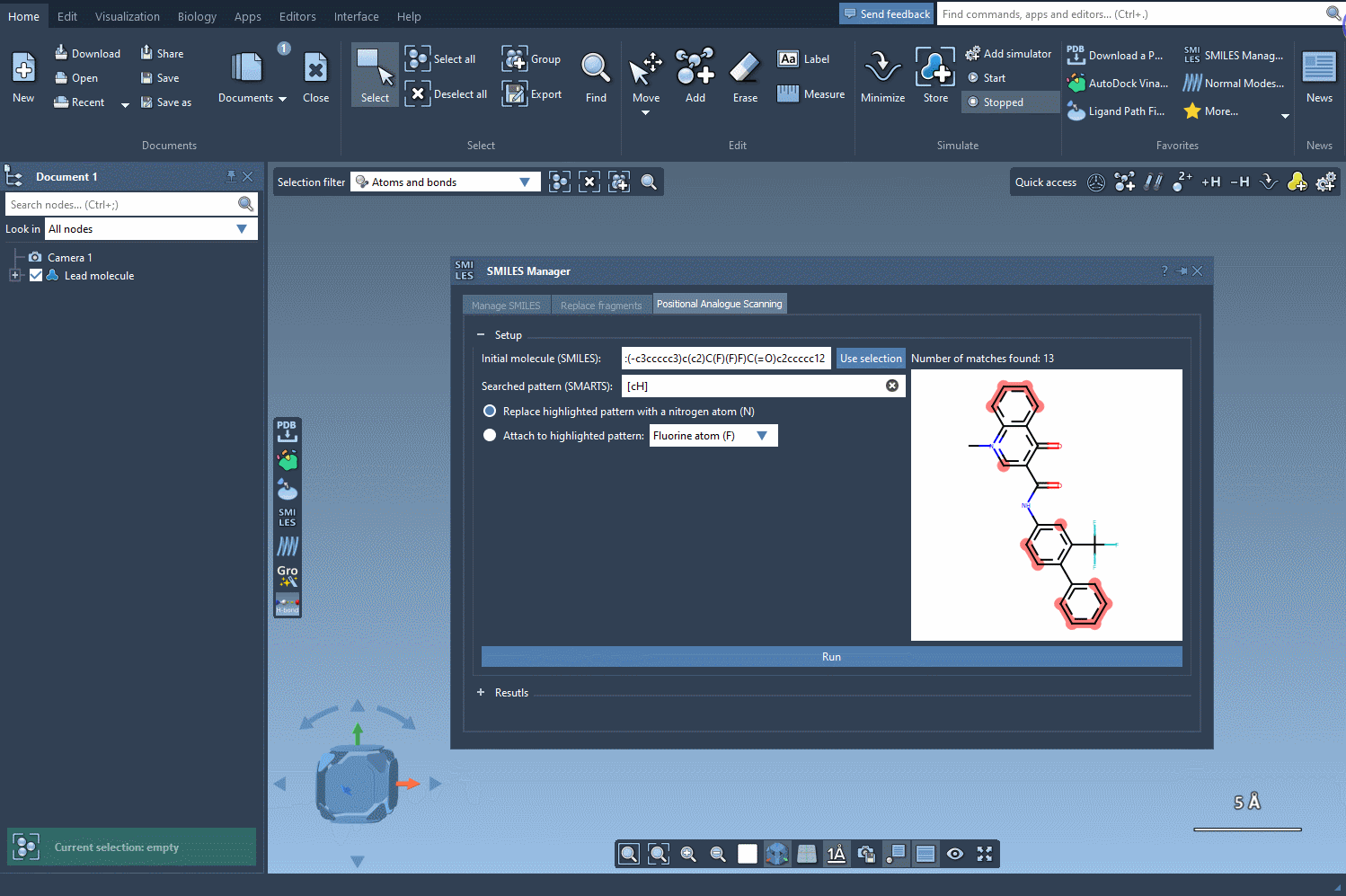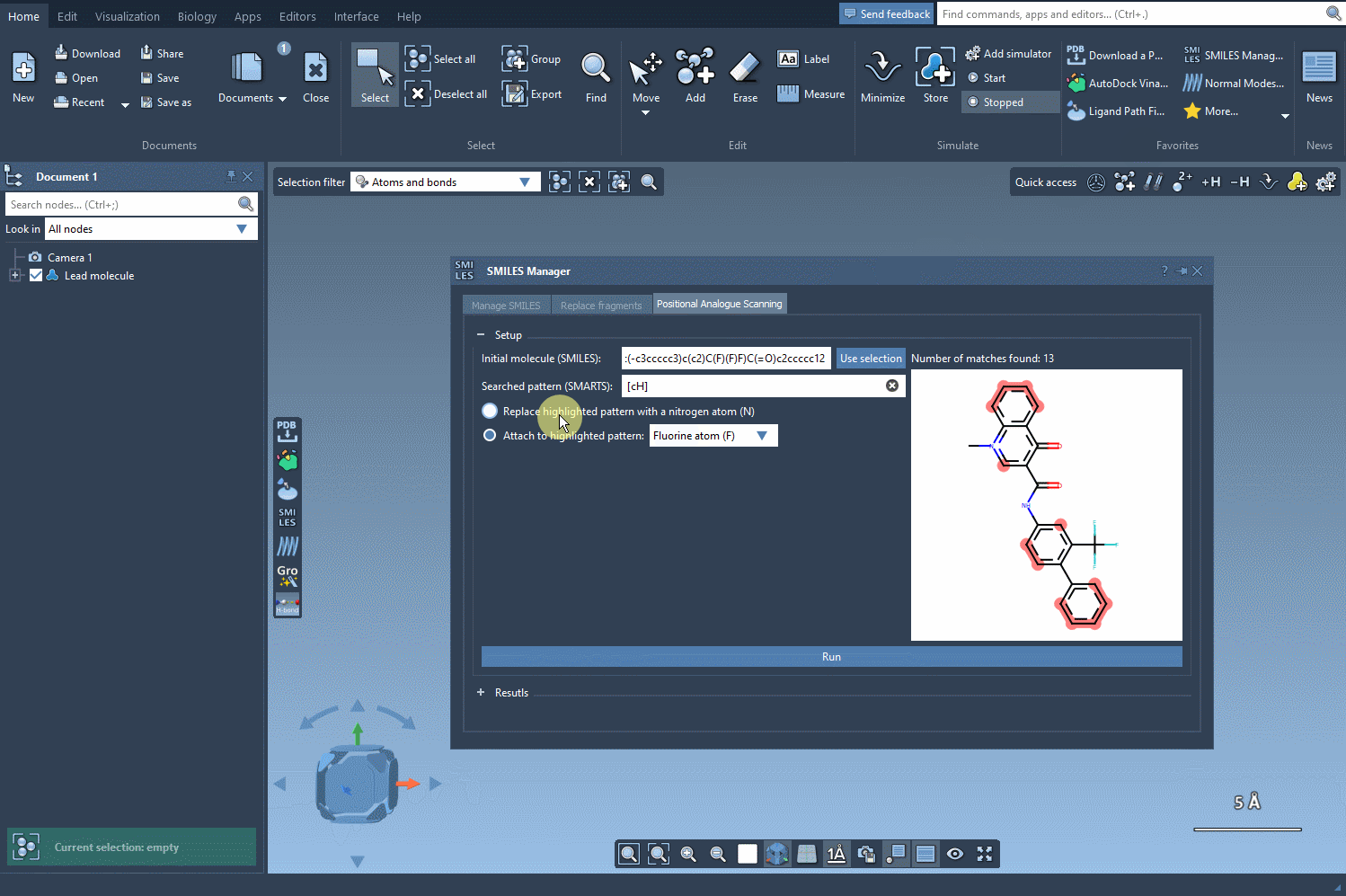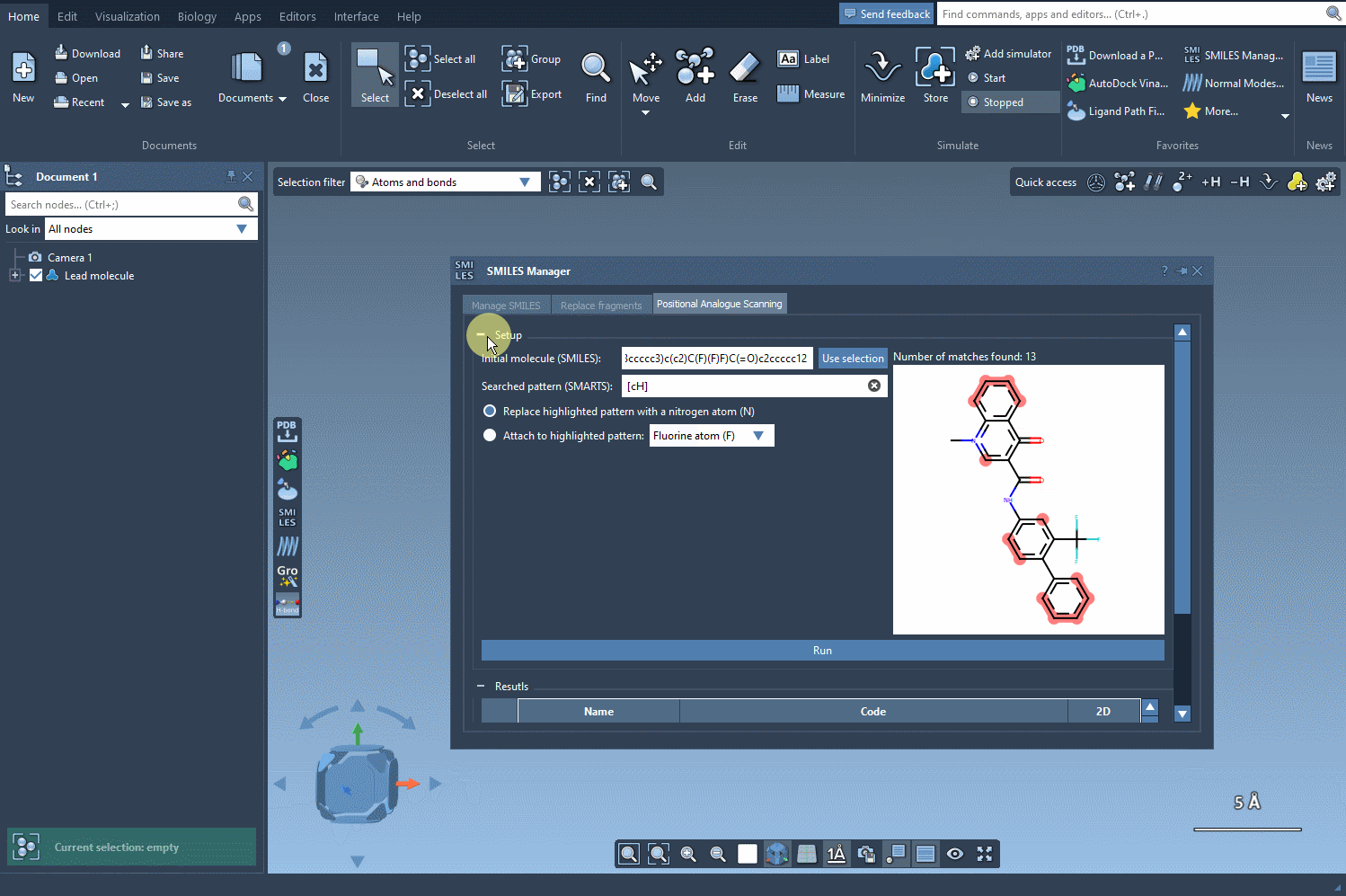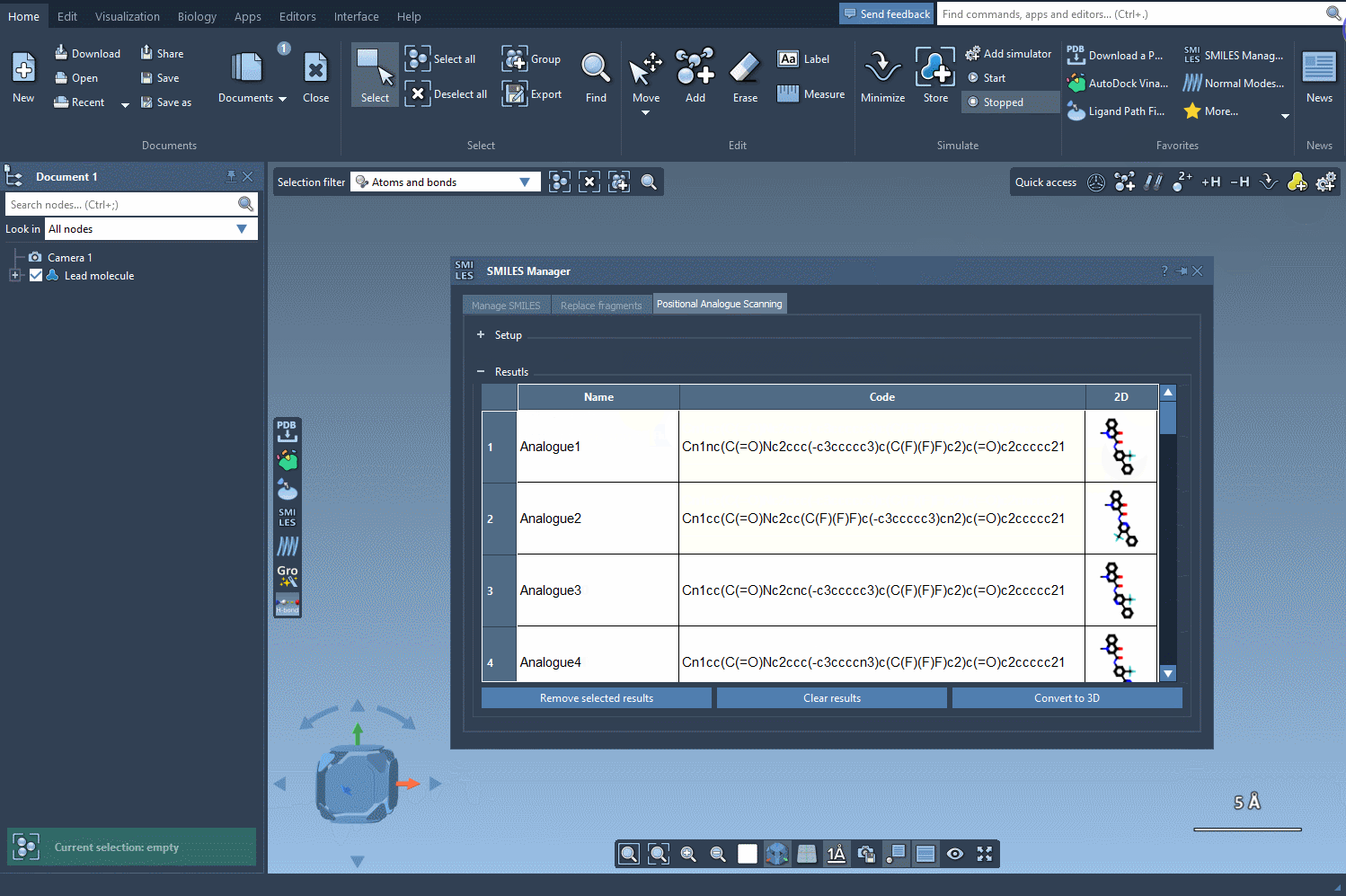
Next, choose your replacement — such as a nitrogen atom (N) — or select an attachment like a fluorine (F) or methyl group (CH3). Click Run, and within seconds, the analogs appear in your results list with their corresponding 2D depictions.
From variation to insight
This workflow frees you to focus on interpretation instead of molecule generation. With your analogs in hand, you can:
- Edit their names or SMILES codes directly through the interface
- Visualize the 2D depictions or enlarge them for clarity
- Remove unwanted analogs, one by one or in bulk
Better yet, if a particular analog looks promising, generate its 3D structure with a couple of clicks and run a docking simulation using the Autodock Vina Extended extension to inspect interactions within a binding site.
Conclusion
Whether you’re early in your design cycle or evaluating lead optimization strategies, the ability to easily replace a molecular pattern and explore analog series in SAMSON can reveal patterns and insights you may otherwise overlook.
To learn more, visit the full documentation page on positional analogue scanning with the SMILES Manager: Read the tutorial.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





