When working with molecular models, one frequent challenge scientists face is modifying structures without breaking the simulation pipeline. Whether you want to prototype a new molecule, adjust a ligand, or refine a complex configuration, editing molecular bonds can easily disrupt your topology and force you to restart. What if this process could be smoother, more forgiving, and physically guided?
The Interactive Modeling Universal Force Field (IM-UFF) in SAMSON offers a solution by allowing dynamic topological changes directly during manipulation. Let’s explore how this works and why it can save time in your modeling workflows.
Why interactive modeling matters
Traditional force fields like UFF are powerful for simulations, but they often assume a fixed topology — set bonds, unchangeable atom types, and predefined connectivity. This works well for static models, but breaks down when users want to explore variations on molecular structures.
IM-UFF extends the standard UFF to enable topology changes while modeling. Move an atom too far? Its bond breaks. Bring it close to another one? A new bond may form — smoothly and realistically, all based on energy criteria.
How it works in practice
- Open your molecular system in SAMSON.
- Add a simulator via Edit > Simulate > Add simulator (or press
Ctrl + Shift + M/Cmd + Shift + Mon macOS). - Select the Interactive Modeling Universal Force Field from the interaction model list.
- Choose a state updater (e.g., FIRE).
- Start the simulation by selecting Edit > Simulate > Start.
Now try dragging an atom with your mouse. If you move it gently, the structure just flexes. If you drag it far enough, bonds break naturally. And when you bring it closer to another atom, new bonds may form. This creates an intuitive and interactive experience, backed by physically meaningful rules.
Customization options
IM-UFF comes with two specific settings that affect real-time editing:
- Static topology (UFF only): When checked, disables dynamic changes (you’re using standard UFF). When unchecked, you’re in interactive mode with IM-UFF.
- Keep vdW for manipulated: Controls whether van der Waals forces are applied to atoms being moved by the mouse. Turning it off may help prevent repulsion from blocking connections when forming new bonds.
You’ll find these options in the IM-UFF parameter panel after starting the simulation — intuitive and accessible.
Real-time editing in action
Here’s an example of what it looks like when you’re editing a molecule with IM-UFF enabled:
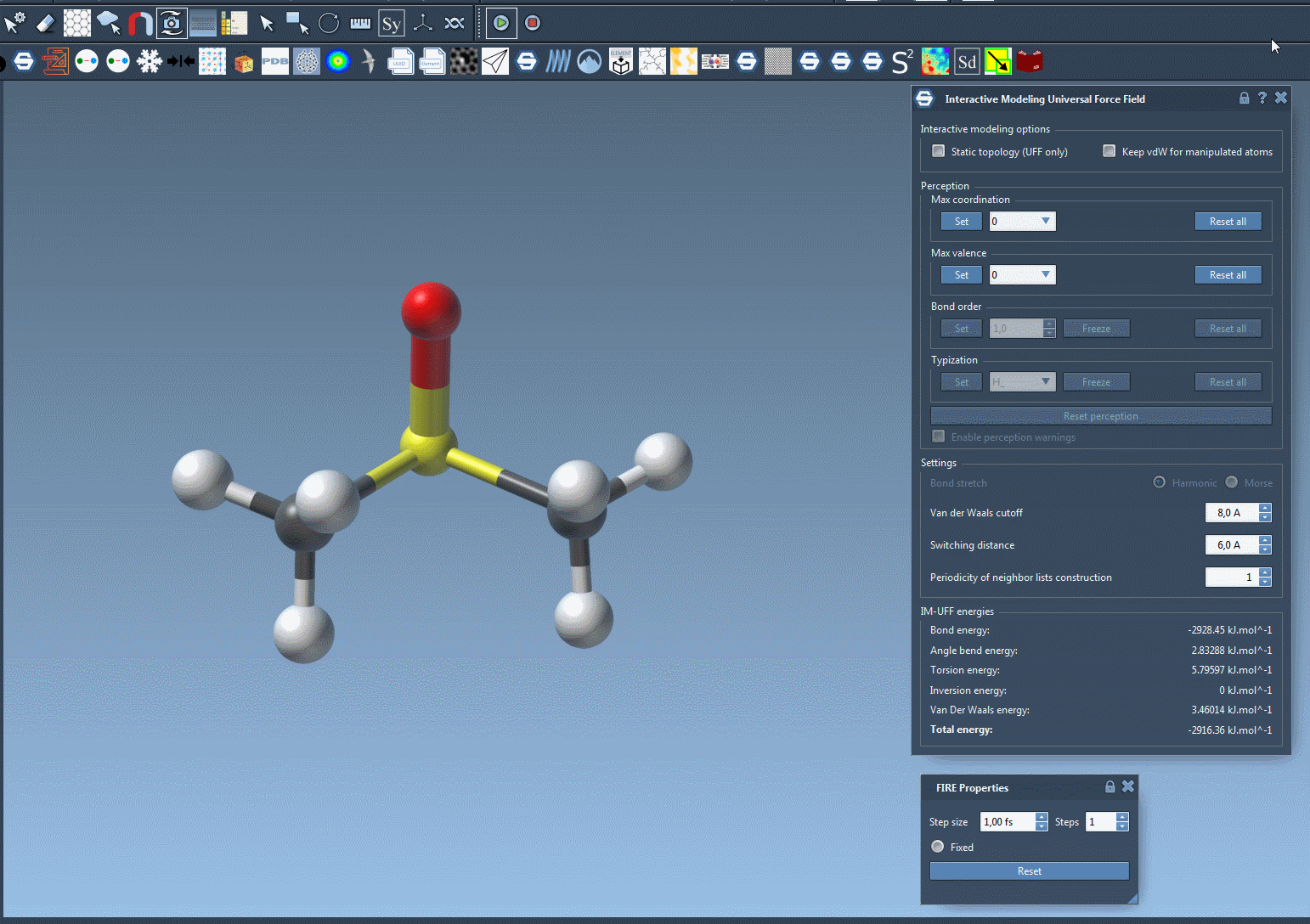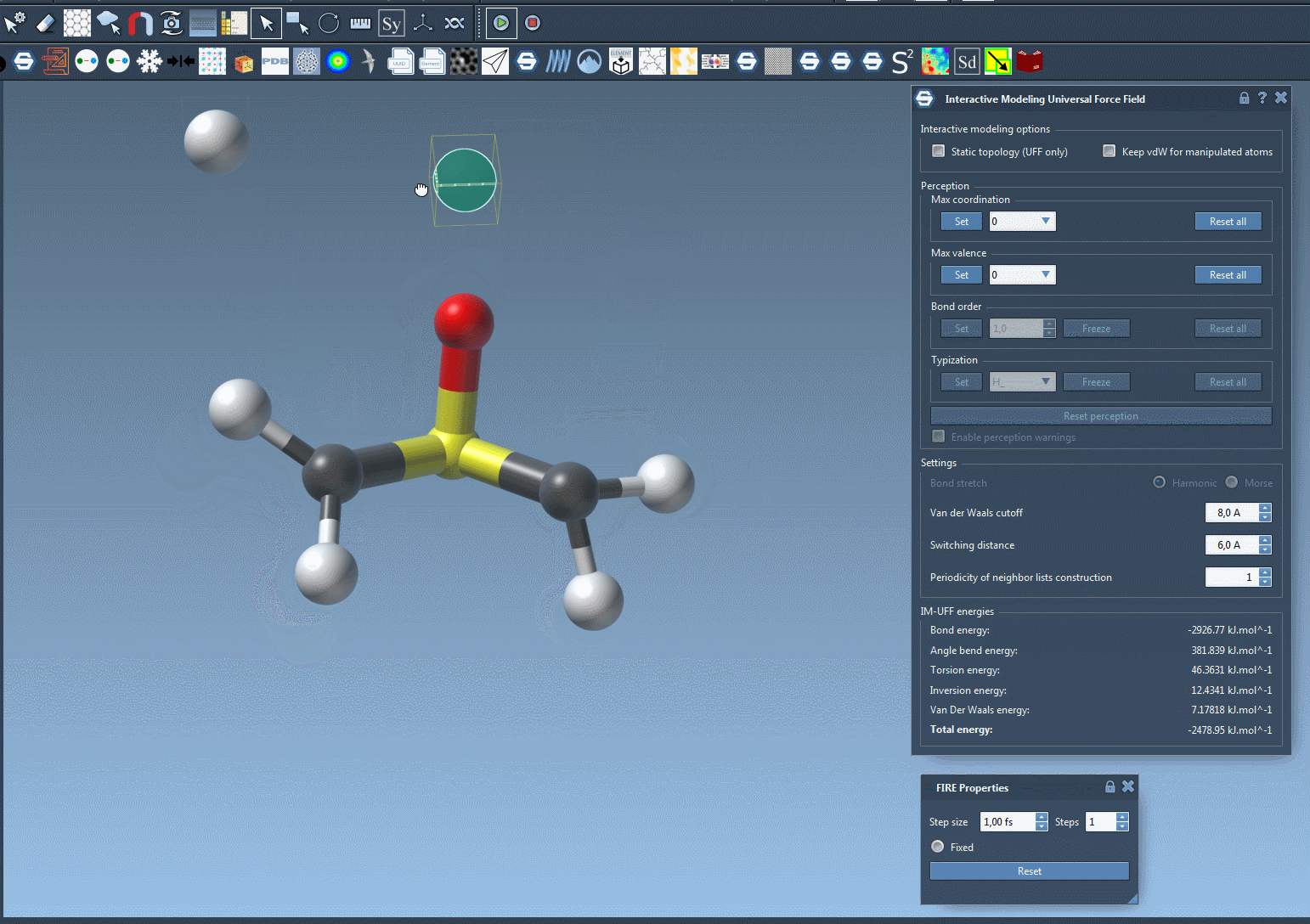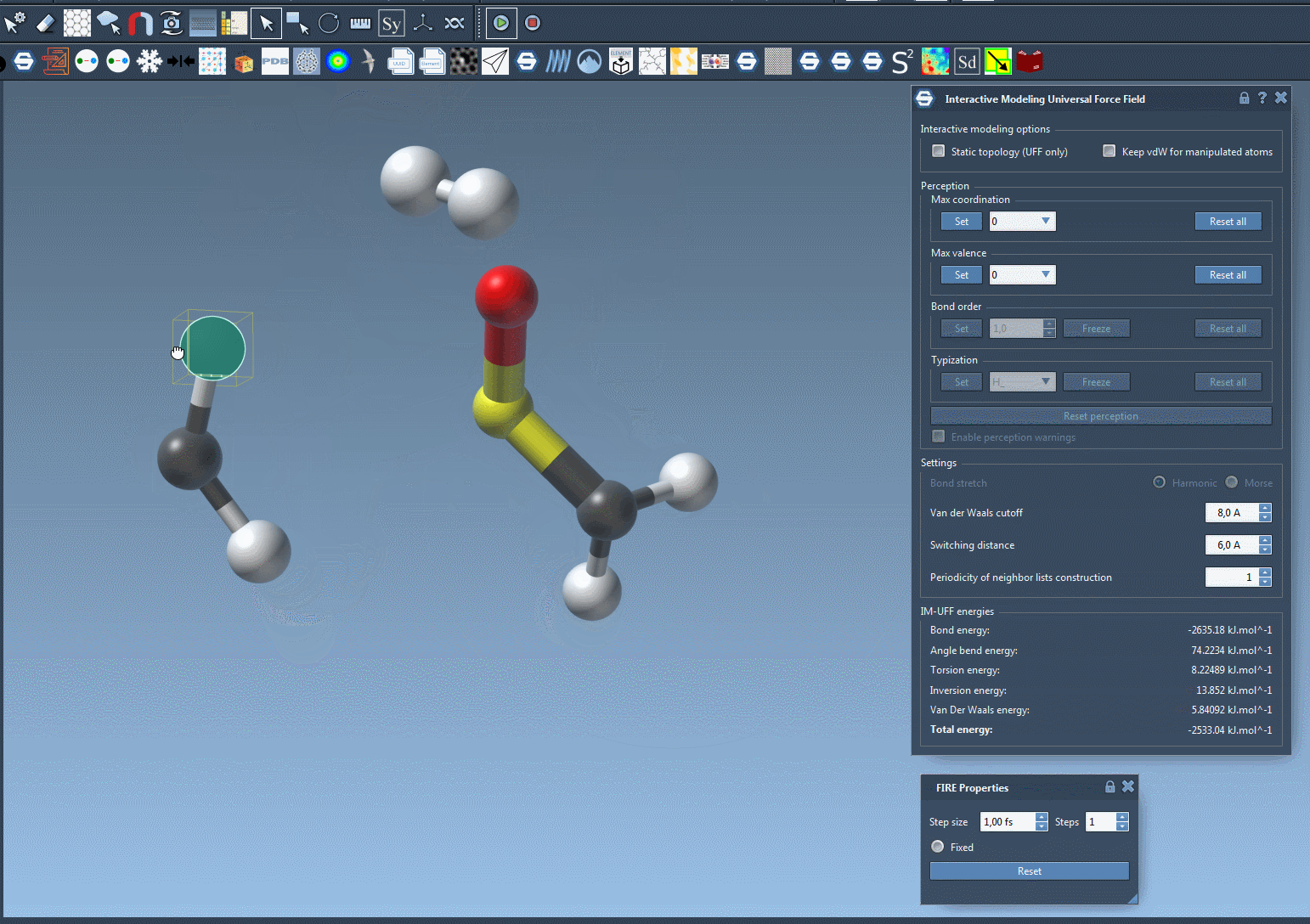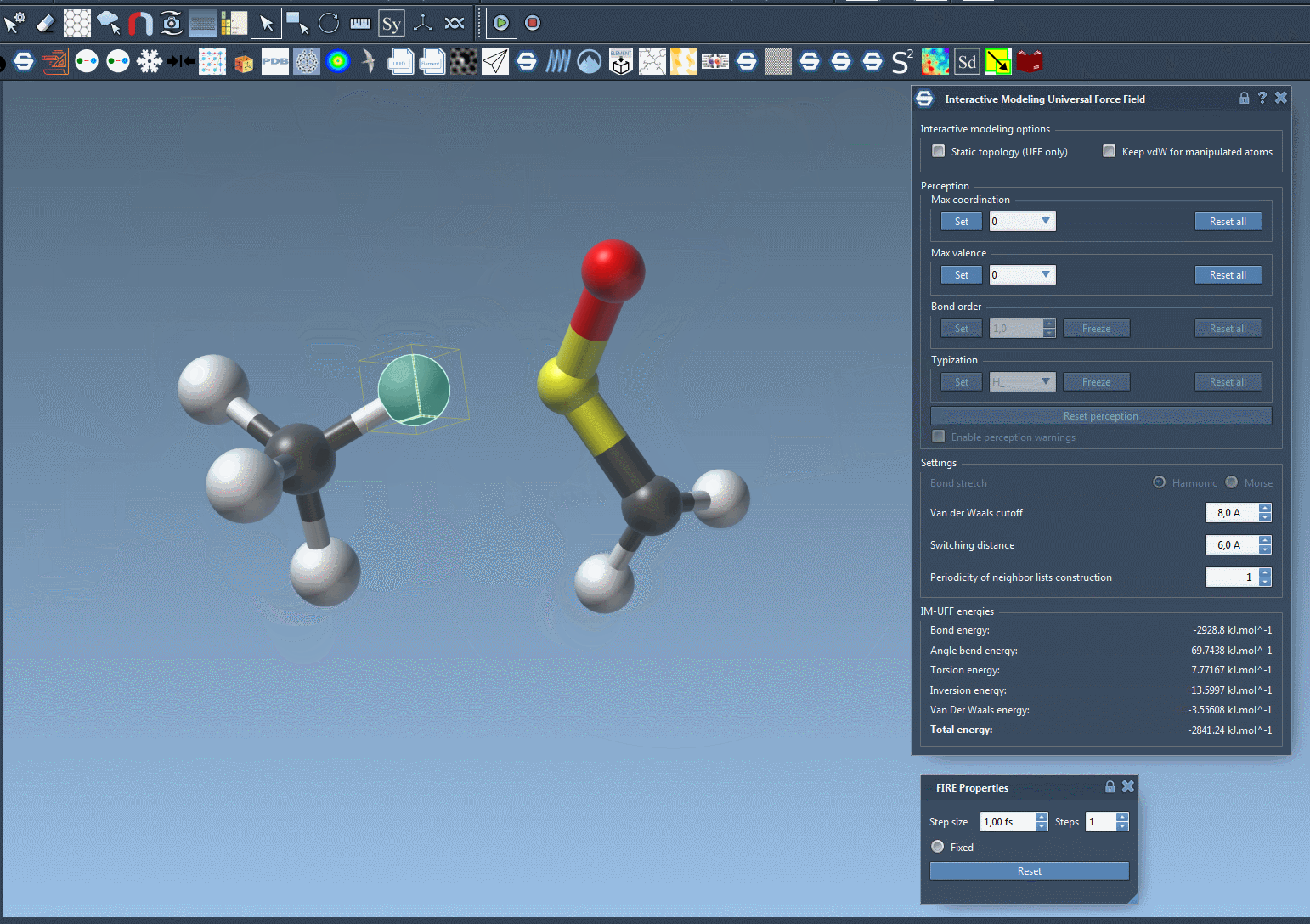
As you manipulate atoms, you’ll notice how SAMSON smoothly updates molecular structures. Topological edits—normally tedious and error-prone—become tractable and efficient.
Who is this for?
If you’re designing new molecules, testing bioactive conformations, or experimenting with chemical interactions, dynamic bond updates help you iterate faster. You don’t have to stop the simulation, go back to editing mode, compute topology, then restart.
Instead, you explore molecular changes on the fly, supported by a force field that updates atom types and bond orders based on geometry and coordination.
To learn more about how this works and how to customize various parameters, visit the full IM-UFF documentation here:
https://documentation.samson-connect.net/tutorials/uff/im-uff/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





