One of the most common challenges in molecular modeling today is generating reliable 3D structures of protein-ligand complexes when experimental data isn’t available. Whether you’re trying to evaluate a drug candidate, explore a new biomolecular interaction, or prepare input files for simulations, having accurate structural models is essential.
Using the Boltz-2 service within SAMSON’s Biomolecular Structure Prediction extension can help address this issue. In just a few steps, you can generate realistic structural models of single molecules or complexes that include proteins, nucleic acids, and small molecules with optional modifications. If you’ve ever faced the frustration of hunting for structures in public databases and still being left empty-handed, read on.
Why Use Boltz-2?
Boltz-2 is designed to offer flexibility while maintaining ease of use. It’s particularly useful if you:
- Work with biomolecular systems that are not available in the Protein Data Bank (PDB).
- Need to preview and share interaction models like protein-ligand complexes before committing resources to simulations.
- Want to test out experimental modifications or design novel biomolecular systems quickly.
How to Start a Prediction with Boltz-2
- Navigate to Home > Predict in SAMSON.
- Select Boltz-2 from the prediction service list.
- Click Add protein, Add DNA, Add RNA, or Add ligand to input sequences, CCD codes, or SMILES strings. For example:
- For a protein: input the amino acid sequence directly.
- For a ligand: use its SMILES string to ensure the correct chemical structure is captured.
- Need to custom-tailor your molecule? Use the Add modification button to introduce chemical modifications by specifying the residue index and CCD code.
- Once everything is set up, click Start prediction.
Behind the Scenes: Cloud-Powered Predictions
Predictions are carried out on A100 GPUs in the cloud, ensuring reasonable performance even for complex structures. Each job typically costs between 0.5 and 1 computing credit, which you can purchase or request from the SAMSON team if needed.
See It in Action
There’s a five-minute video tutorial that walks you through how to:
- Set up predictions with Boltz-2
- Share results with colleagues
- Generate protein-ligand interaction diagrams
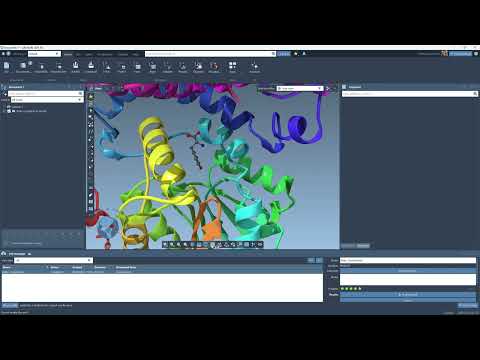
What Happens After Prediction?
Once generated, your models can be accessed from Interface > Cloud jobs or directly on your SAMSON Connect account. Structures loaded into SAMSON may be automatically colorized based on predicted confidence values if such metrics (like pLDDT scores) are included.
Is This For You?
If you’re doing structure-based drug design, building models for molecular simulations, or simply exploring biomolecular hypotheses, Boltz-2 offers an efficient way to generate reliable, customizable starting structures.
To learn more about Boltz-2 and SAMSON’s prediction tools, visit the full documentation page at https://documentation.samson-connect.net/tutorials/bsp/bsp/.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





