Designing and optimizing molecular libraries can be a time-consuming and repetitive process, particularly when you need to generate compound variations by swapping out functional groups or fragments. If you’ve ever wanted to explore isosteric replacements or quickly prototype a series of analogues, SAMSON’s SMILES Manager offers a practical feature that’s worth exploring.
Inside the Replace fragments tab of the SMILES Manager extension, researchers can quickly create a library of substituted molecules by specifying a fragment to replace and a set of substitute fragments. This simplifies the synthesis-oriented design phase and integrates nicely with downstream 3D modeling or property analysis workflows.
What is Fragment Replacement?
Fragment replacement is frequently used in medicinal chemistry to explore structural space around a lead compound. For example, swapping a carboxyl group with a sulfonamide can help evaluate stability or bioactivity differences. Traditionally, generating such analogues manually can become tedious, especially for large libraries and repetitive patterns.
Key Steps in SMILES Manager
Here’s how SAMSON simplifies the process:
- Define Initial Molecule:
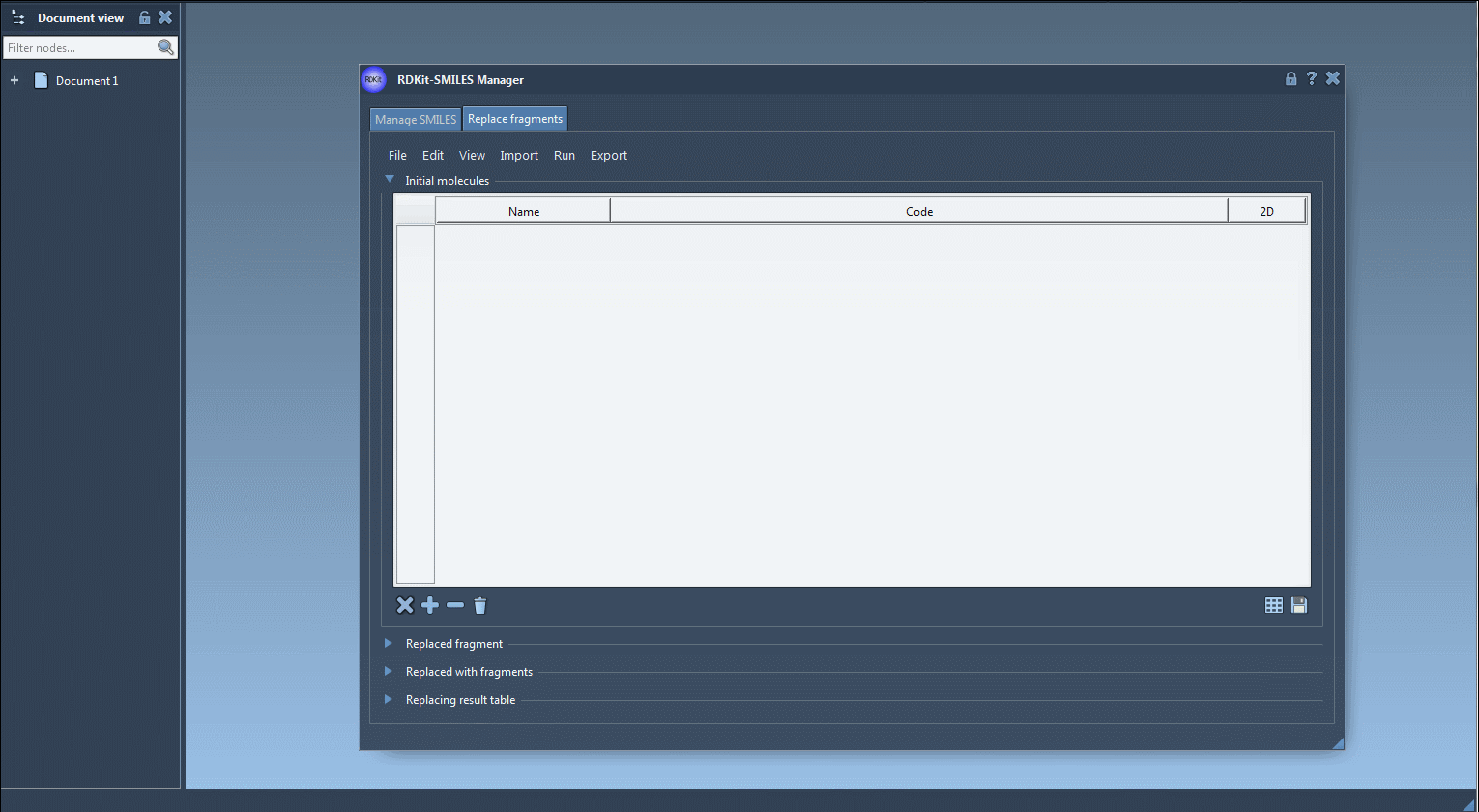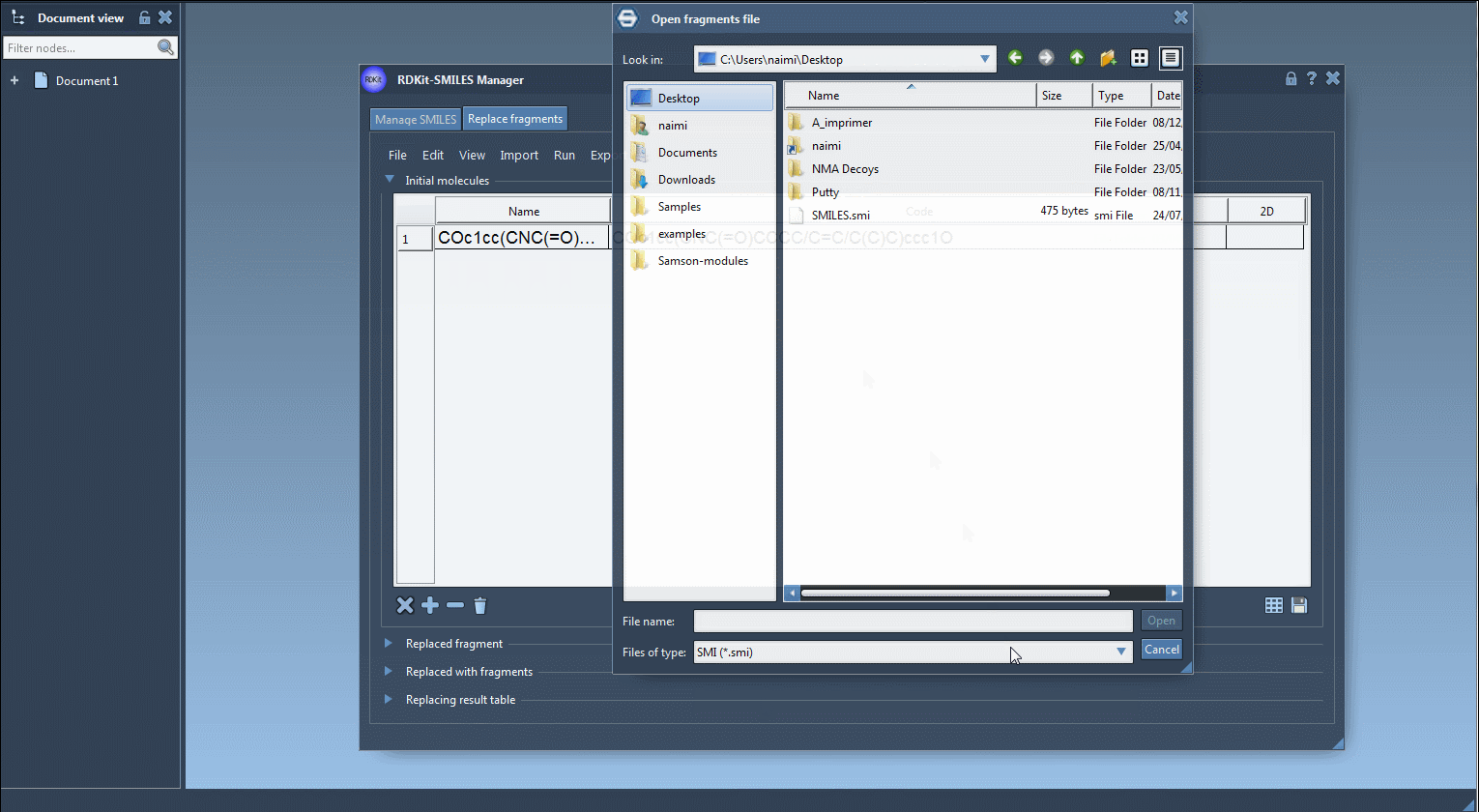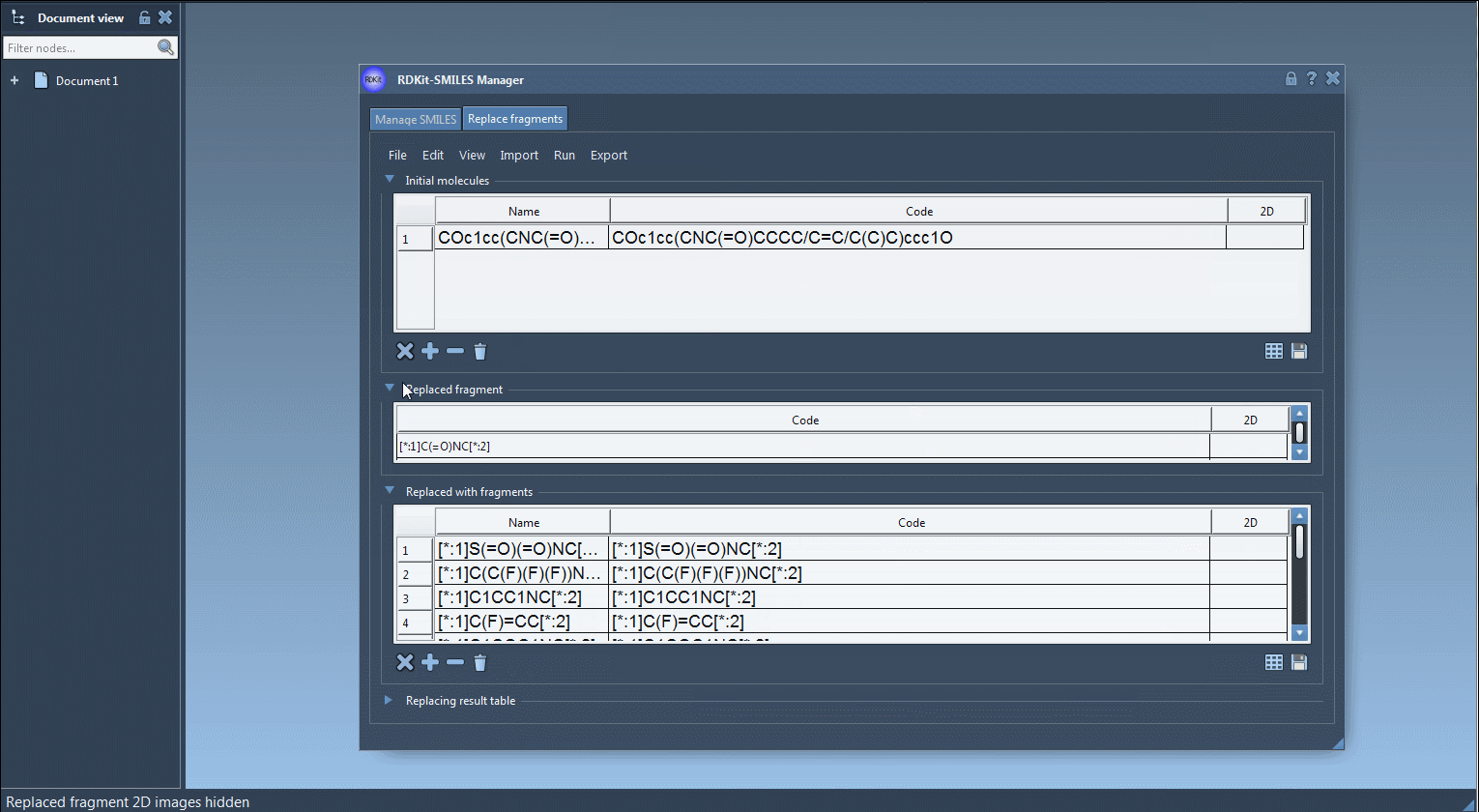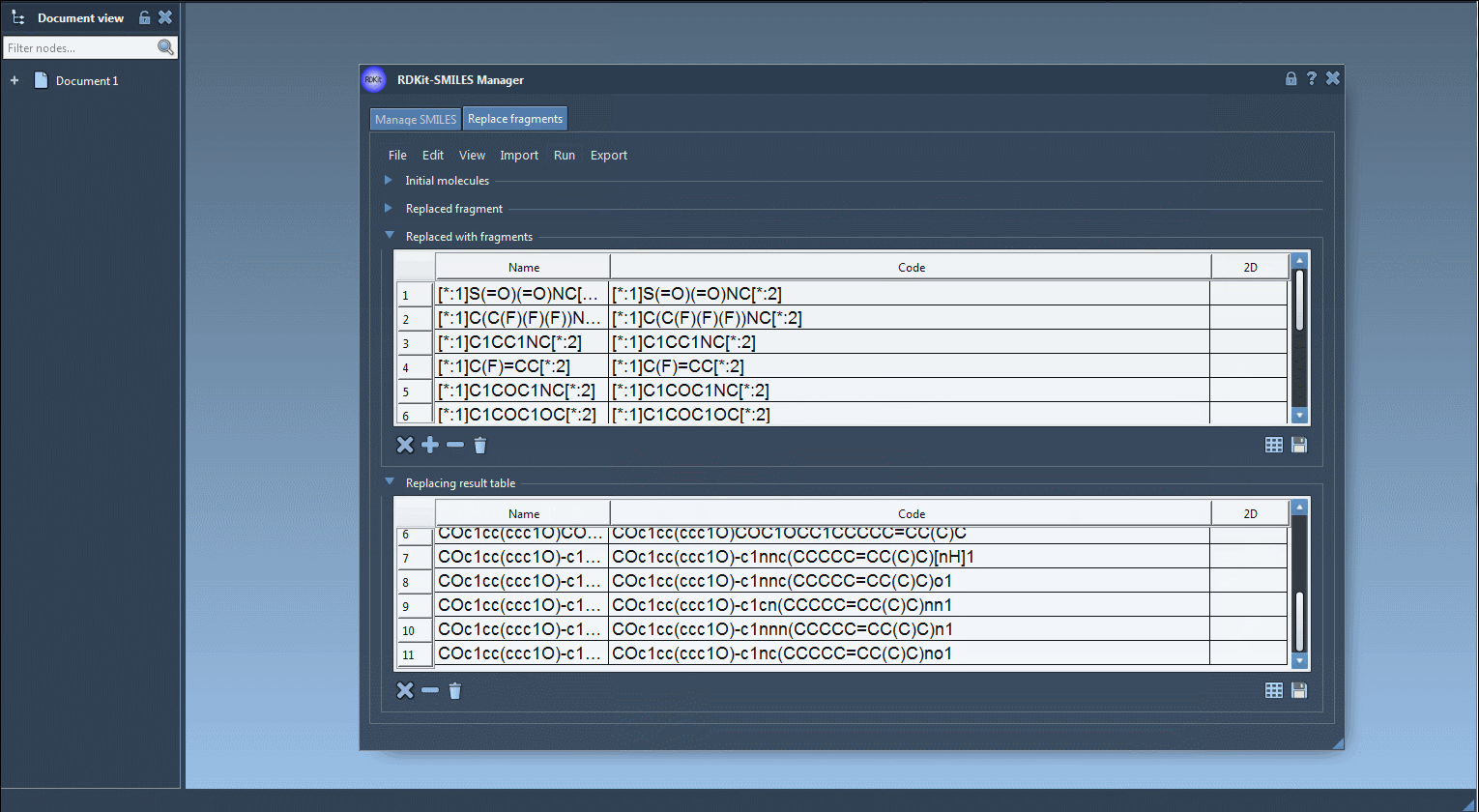
Start by selecting or importing one or multiple initial molecules containing a substructure you want to replace. In the tutorial, the molecule used is:
COc1cc(CNC(=O)CCCC/C=C/C(C)C)ccc1O - Specify the Replaced Fragment:
You can either manually input the SMARTS-like pattern or fetch it from a structure in your active SAMSON document. Example:
[*:1]C(=O)NC[*:2]
This tells RDKit to locate the desired fragment, anchoring on both ends with wildcards. - Provide Replacement Fragments:
Input a list of fragments that the tool will use to generate new molecules. These can be imported from a file or manually entered. For example:
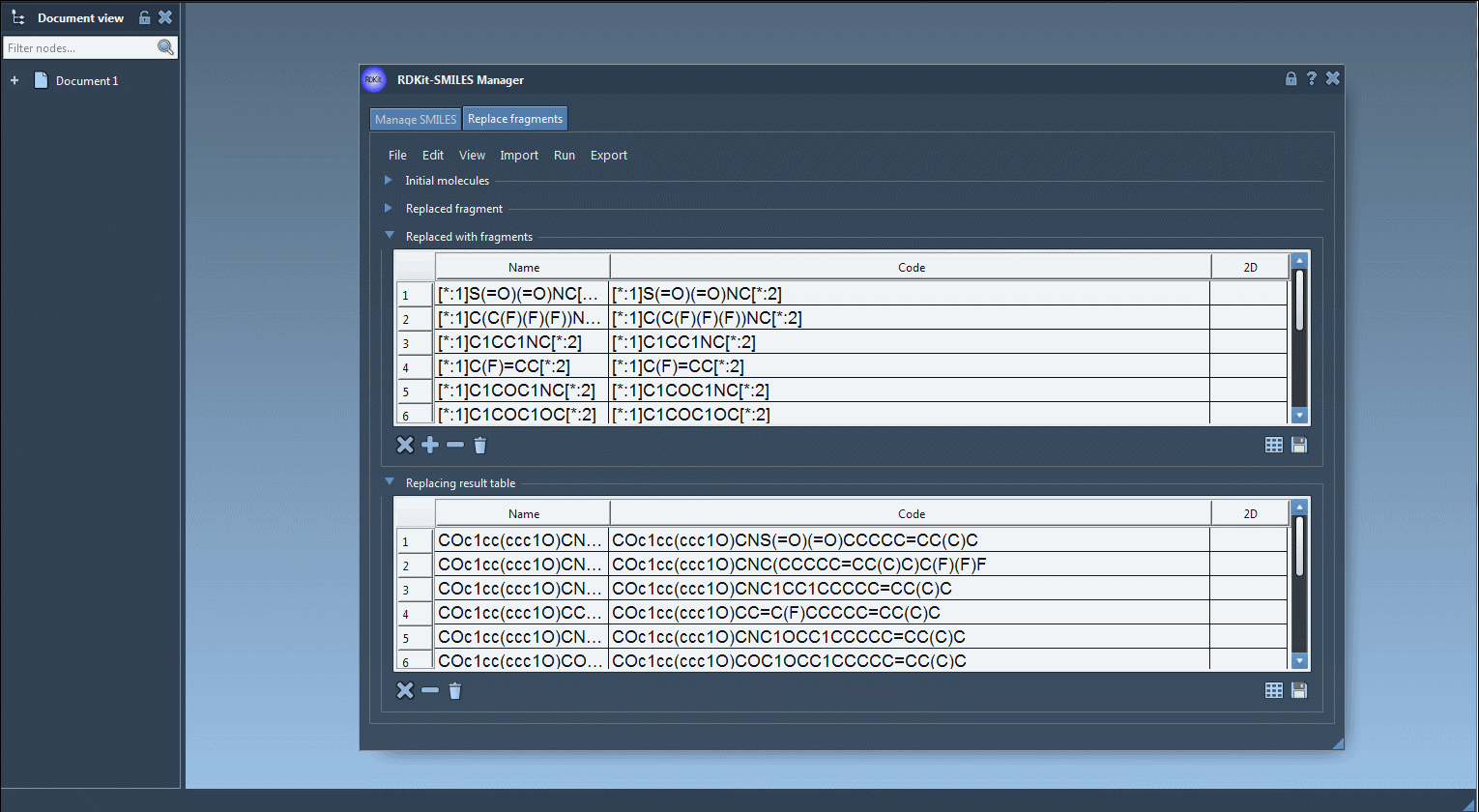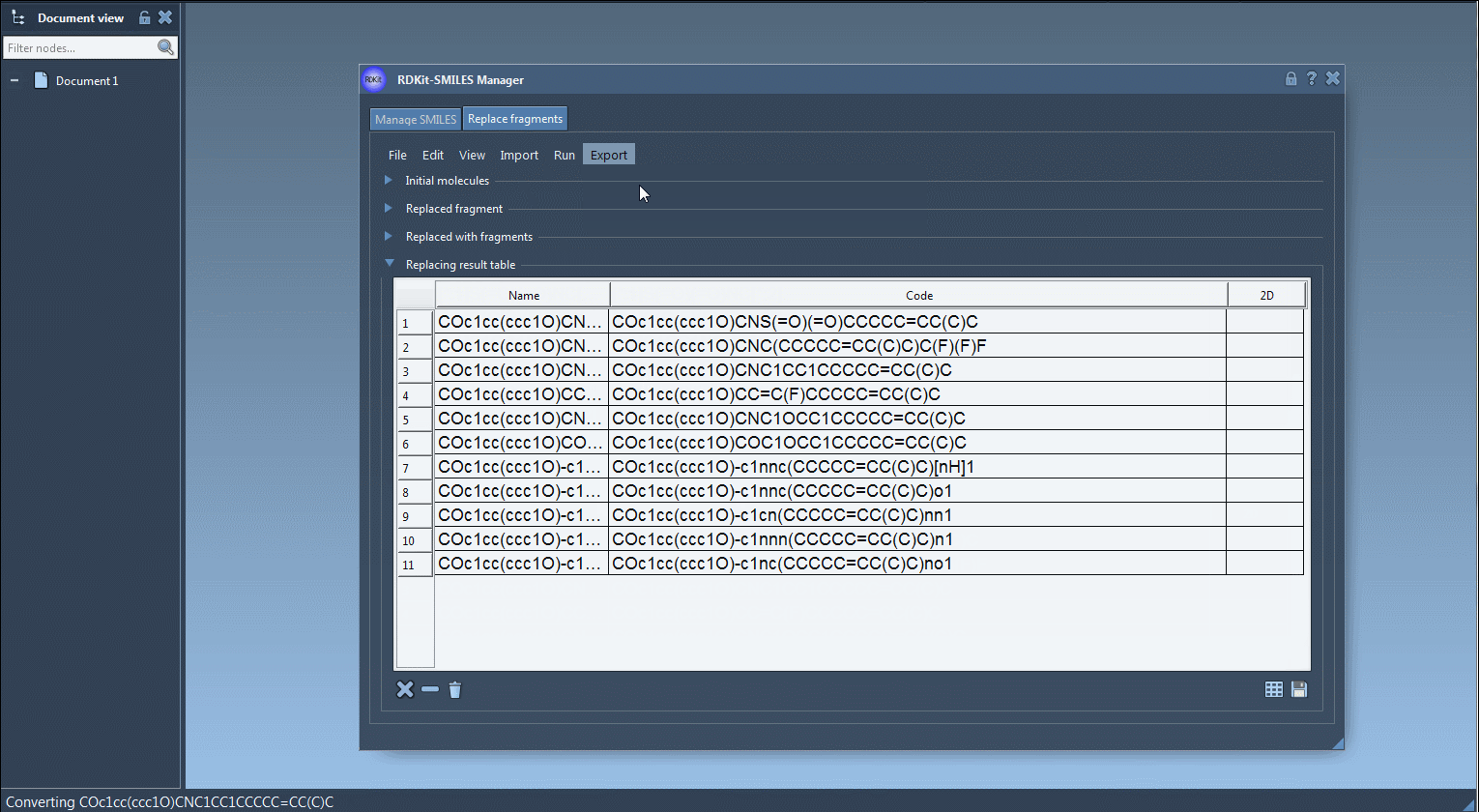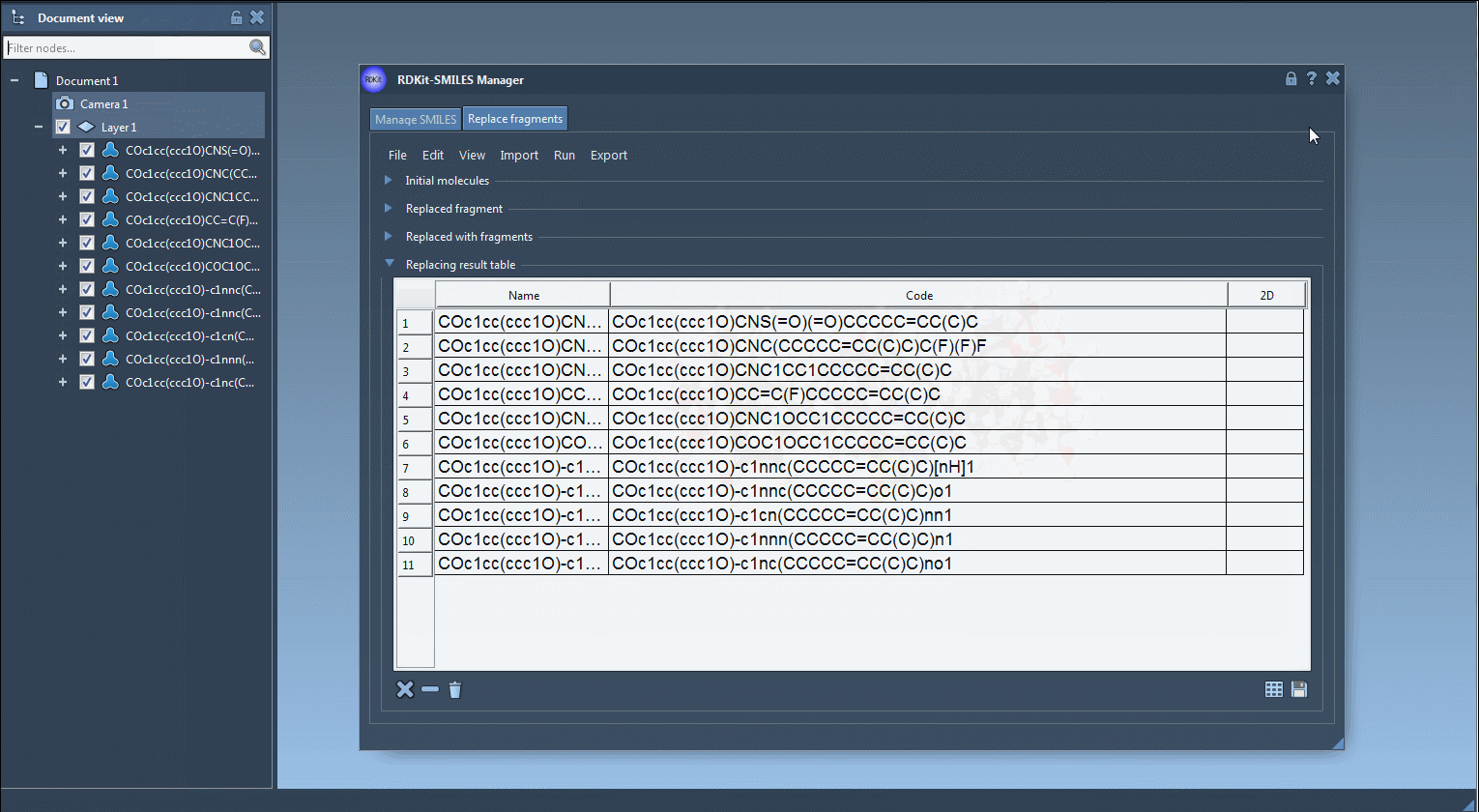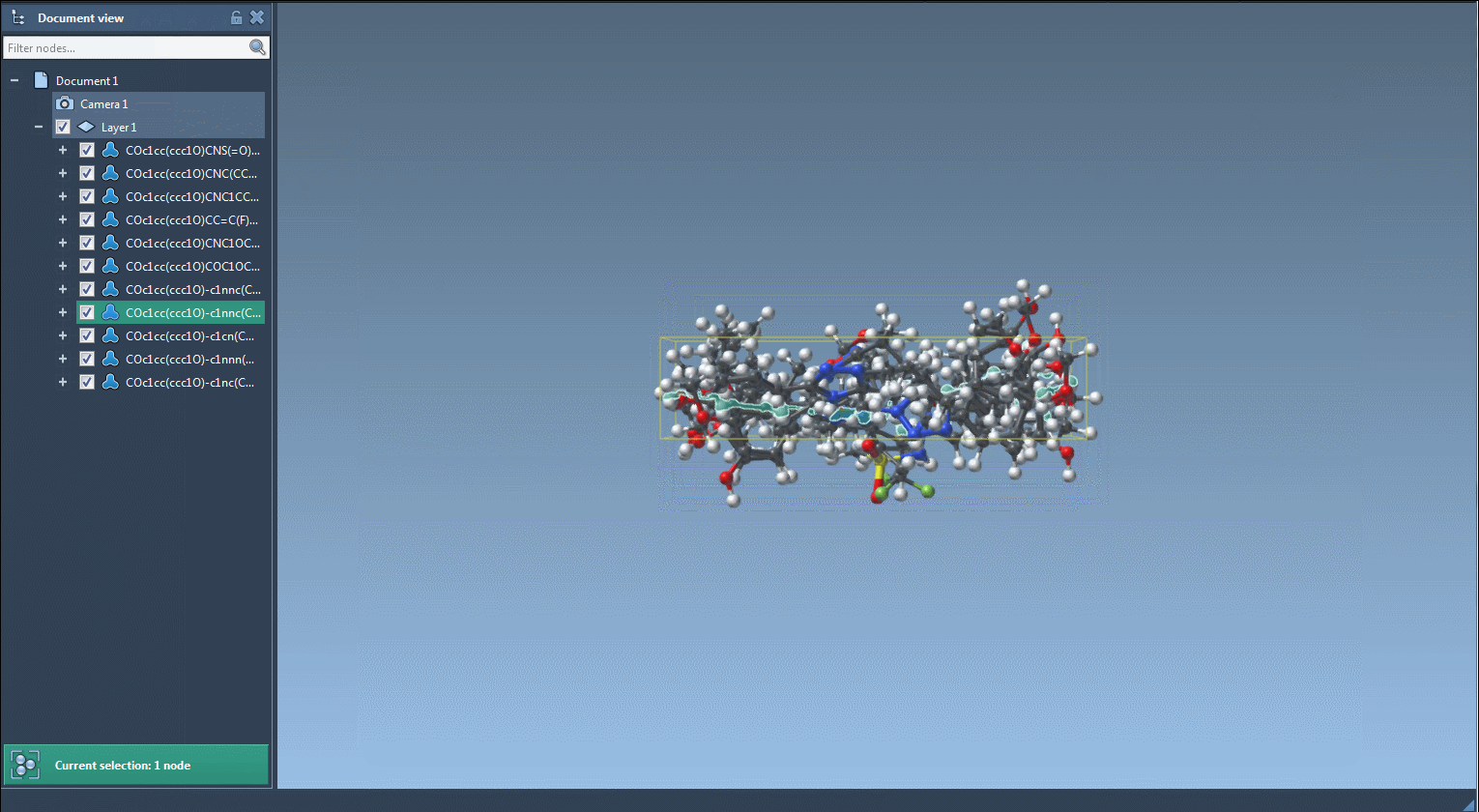
[*:1]S(=O)(=O)NC[*:2] [*:1]C(C(F)(F)(F))NC[*:2] [*:1]C1COC1NC[*:2] ...
Each entry respects the pattern matching needed for consistent replacement and proper bonding. - Run the Replacement:
Click Replace Fragments. The extension automatically generates all variant molecules and displays them in a results table with 2D depictions. - Further Actions:
You can visualize, export, or convert selected variants into full 3D models directly within SAMSON. The results may be saved as individual images or as a grid for documentation or visualization purposes.
Why This Matters
With fragment replacement integrated into SMILES Manager, you can:
- Rapidly build and explore analogue libraries
- Test alternative pharmacophores or side chains
- Generate 3D geometries of variants for docking or simulation workflows
- Avoid repetitive tasks and spend more time analyzing results
Whether you’re performing fragment-based drug design or simply need to test bioisosteric replacements, this capability reduces barrier-to-entry and enhances productivity.
To learn more, check out the full documentation page for the SMILES Manager Extension at https://documentation.samson-connect.net/tutorials/smiles-manager/using-the-rdkit-smiles-manager/.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





