Going from a list of SMILES strings to functional 3D molecular structures can still feel like a chore, especially when you need to process multiple molecules quickly and visualize them in appropriate software. If you’re working in drug discovery, molecular modeling, or any area where molecular representations inform decisions, you’ve likely felt this pain at some point.
This post walks you through using the SMILES Manager extension in SAMSON—a free platform for molecular design—to easily convert multiple SMILES strings into 3D structures using RDKit under the hood. Whether you’re preparing for docking simulations or simply trying to explore chemical space visually, this feature can save you hours of effort.
What This Does So Well
With the SMILES Manager extension, generating 3D molecular structures from SMILES strings becomes an intuitive and transparent process:
- Quickly import SMILES strings from .smi or .txt files
- Visualize these molecules as 2D depictions (for error-checking at a glance)
- Instantly generate and load 3D geometries with just a click
Step-by-Step: From SMILES to 3D
- Import SMILES
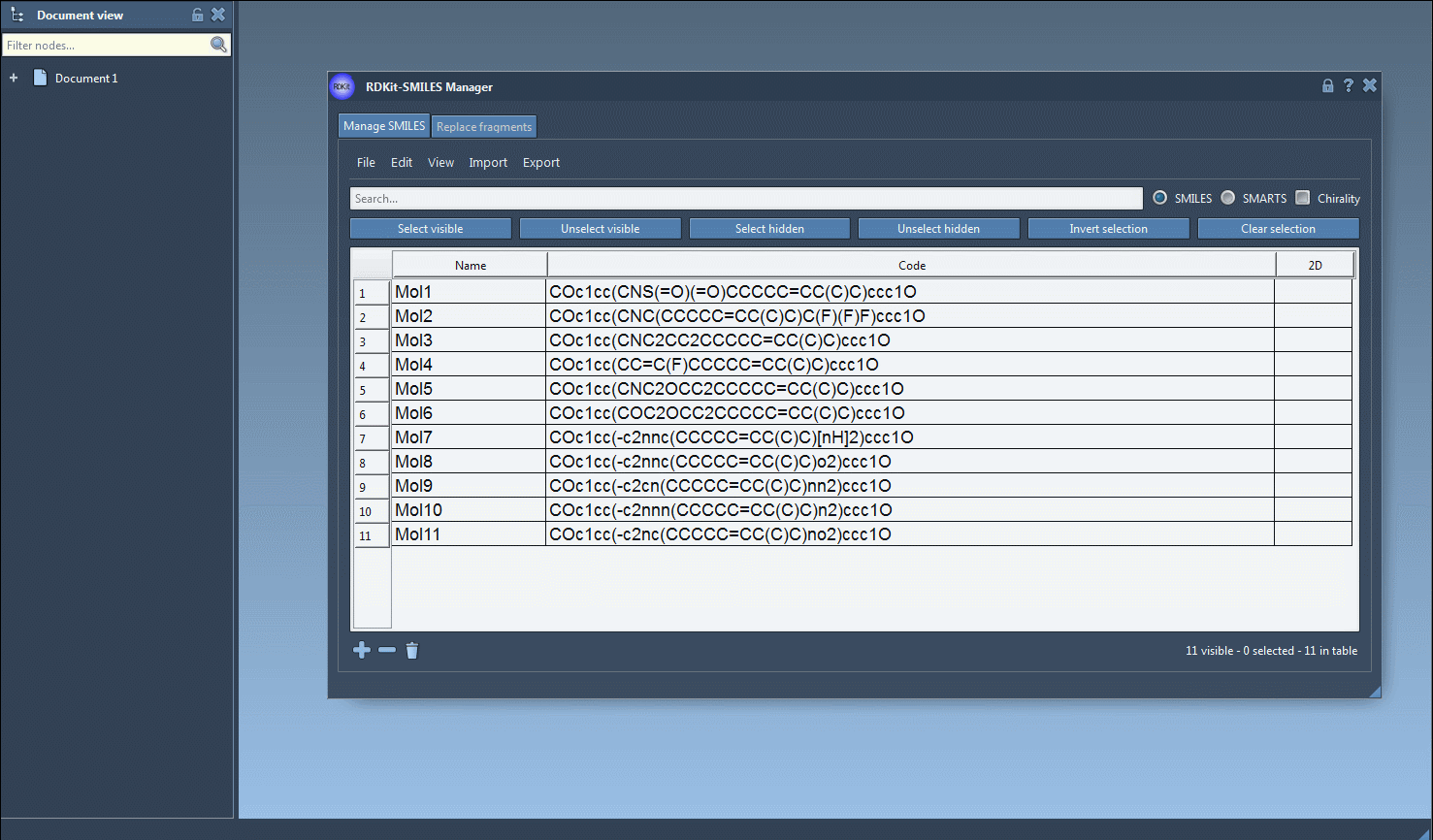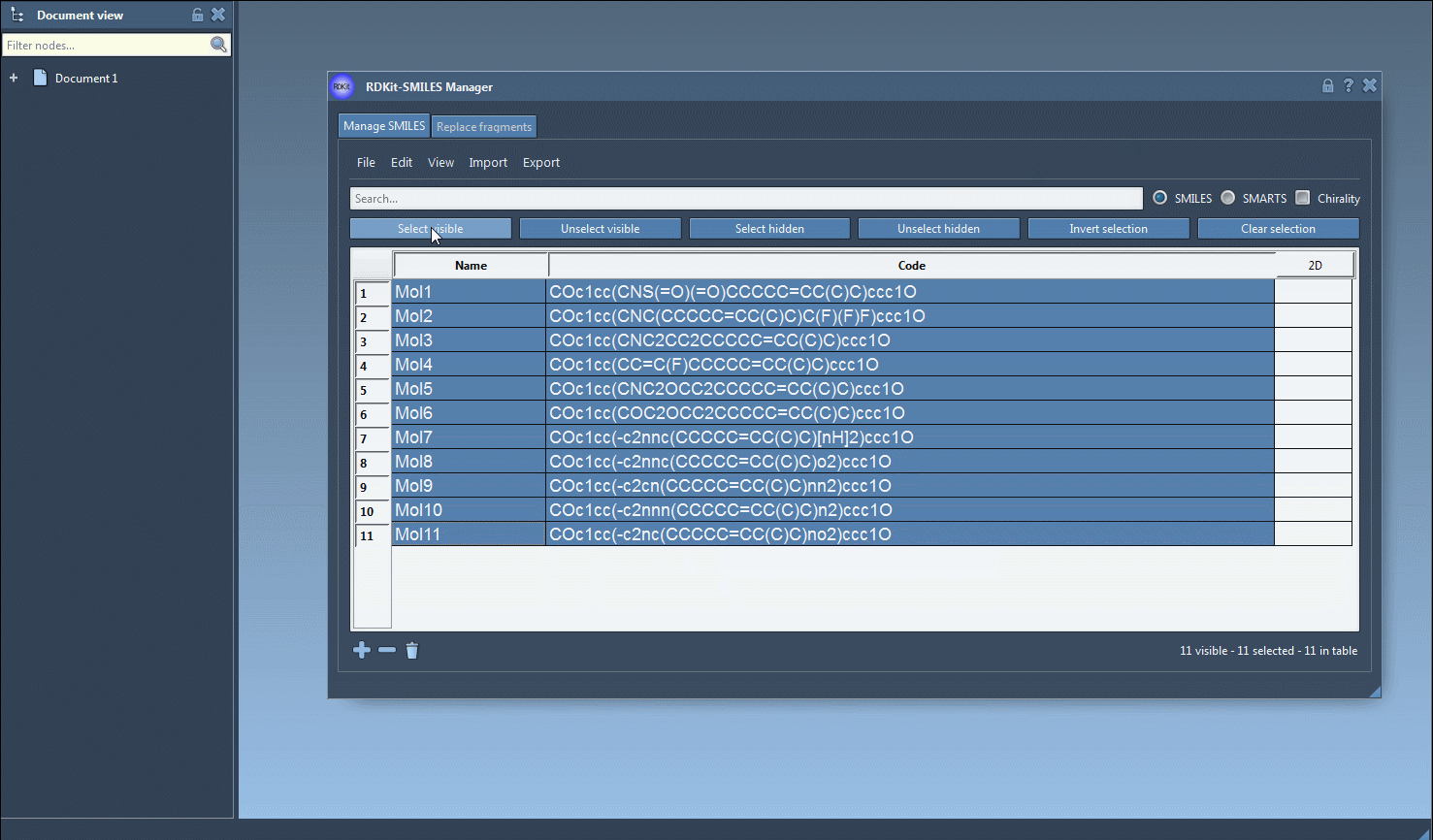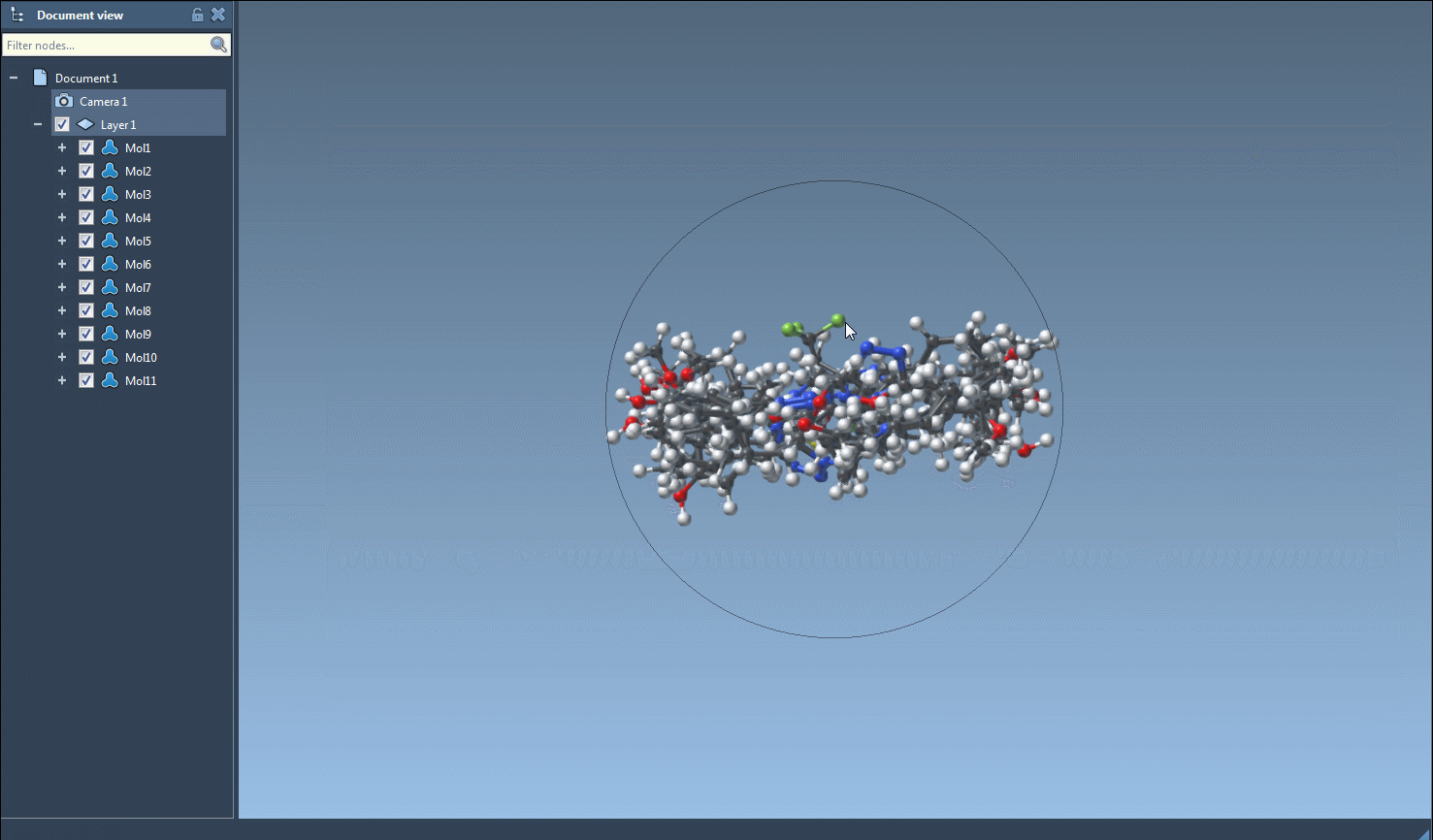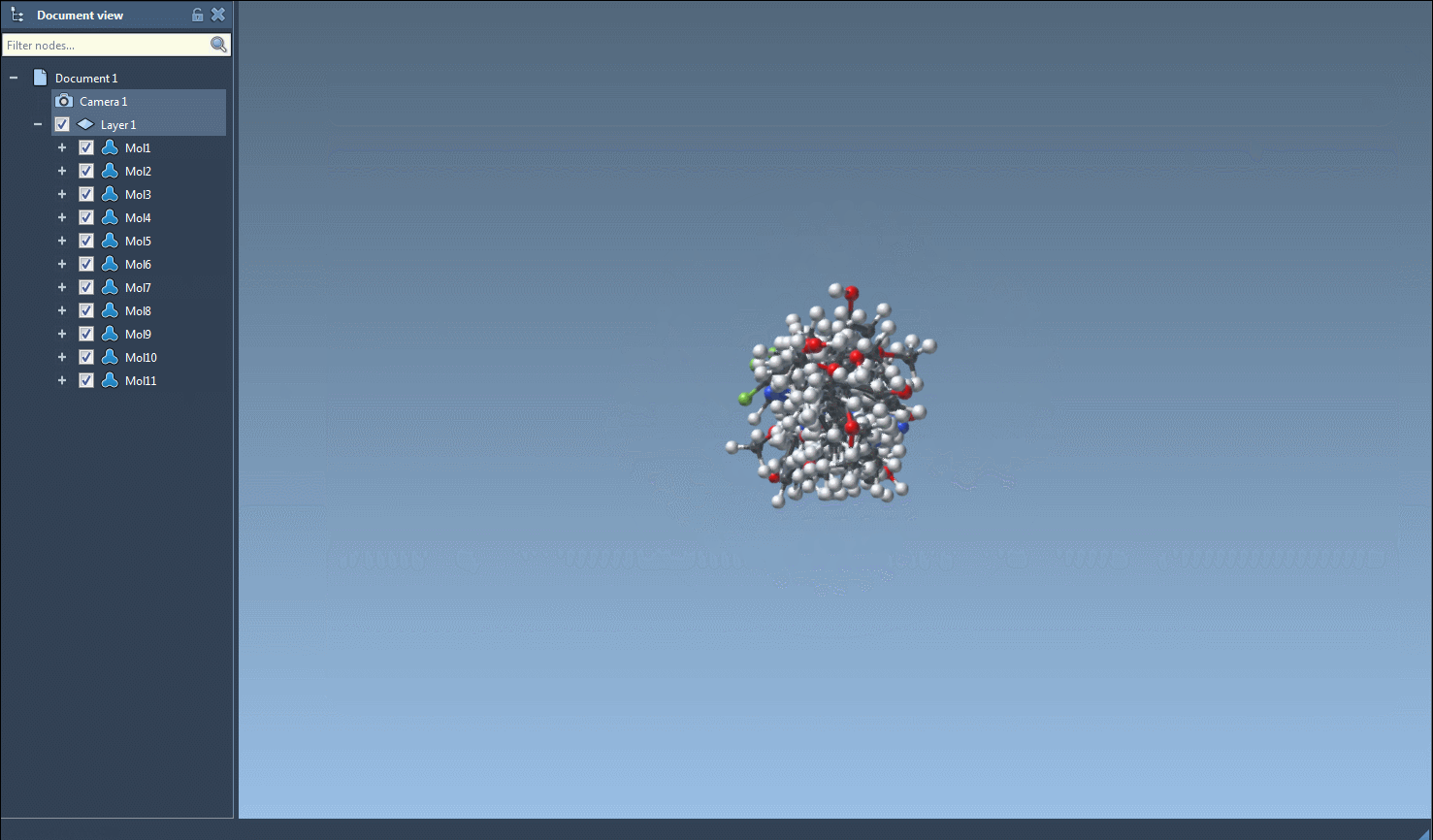
Use the File > Open menu inside the SMILES Manager to load your .smi or .txt files. Each entry contains the SMILES code and an optional molecule name. You can also manually paste or edit within the table.
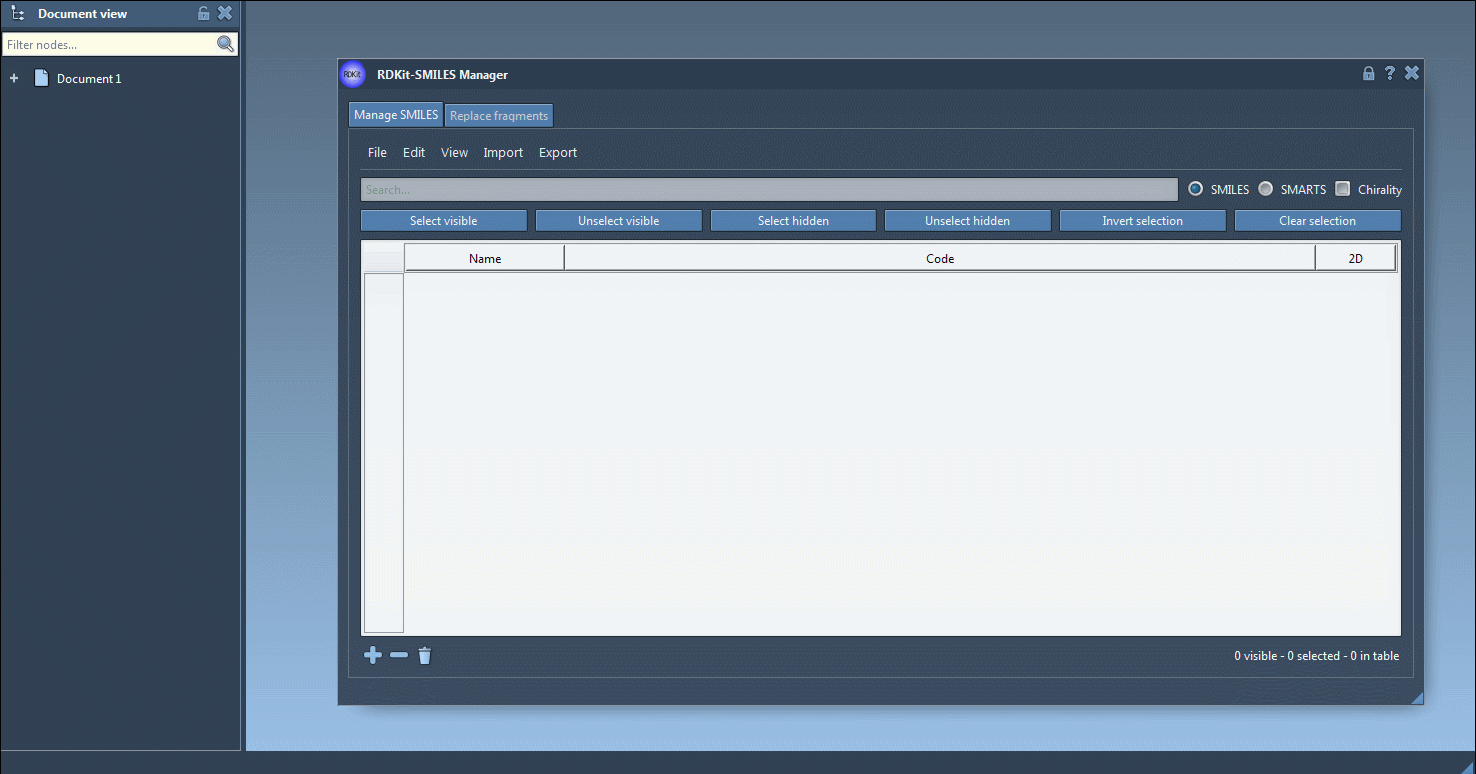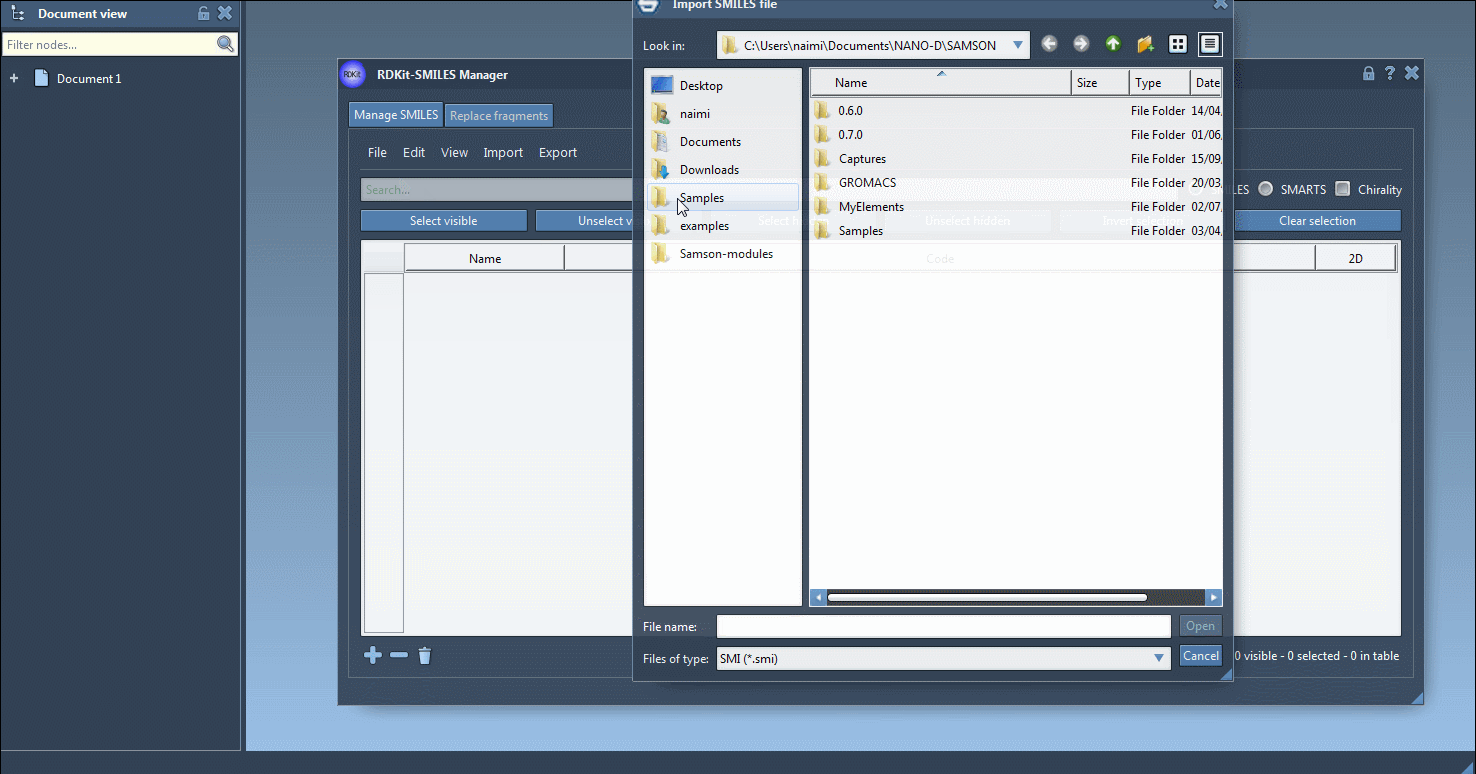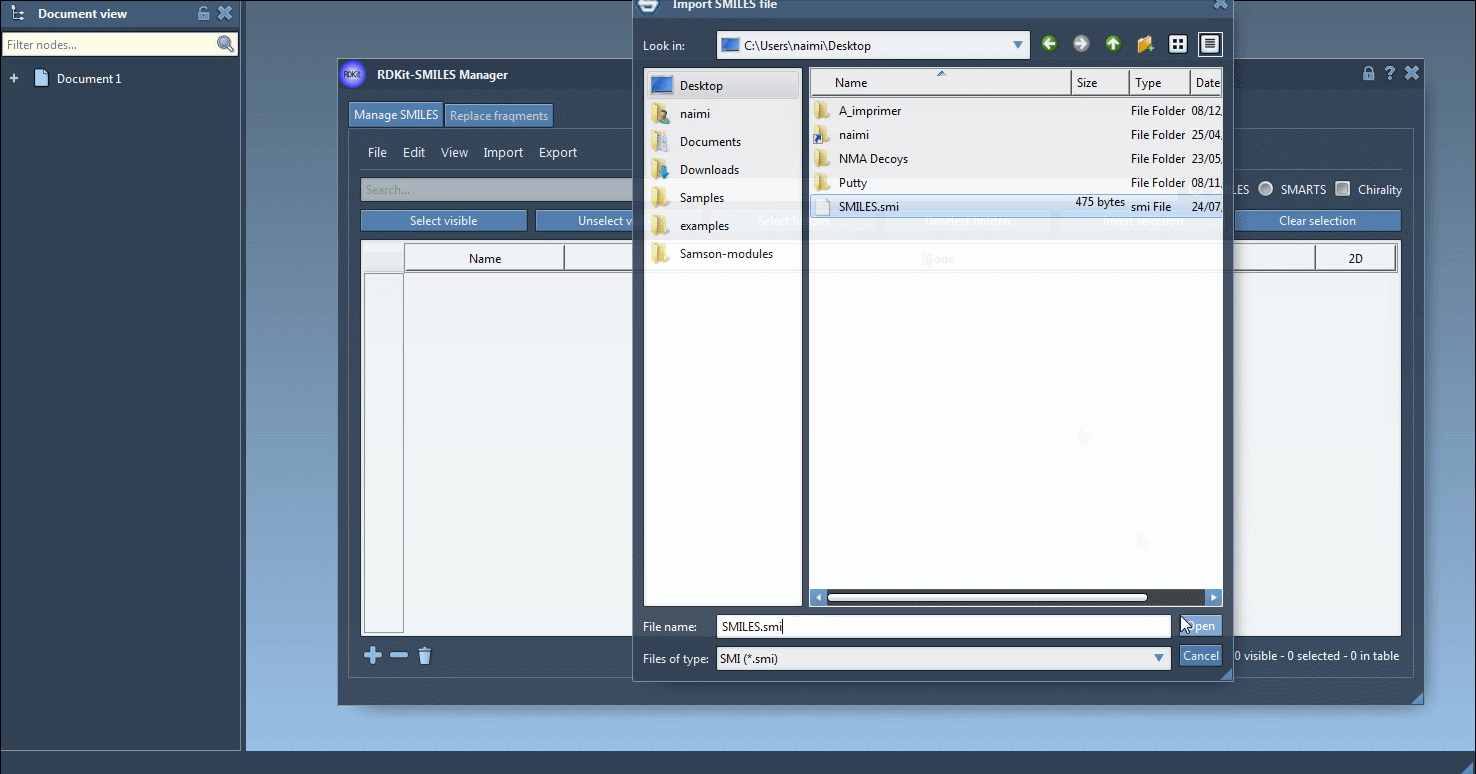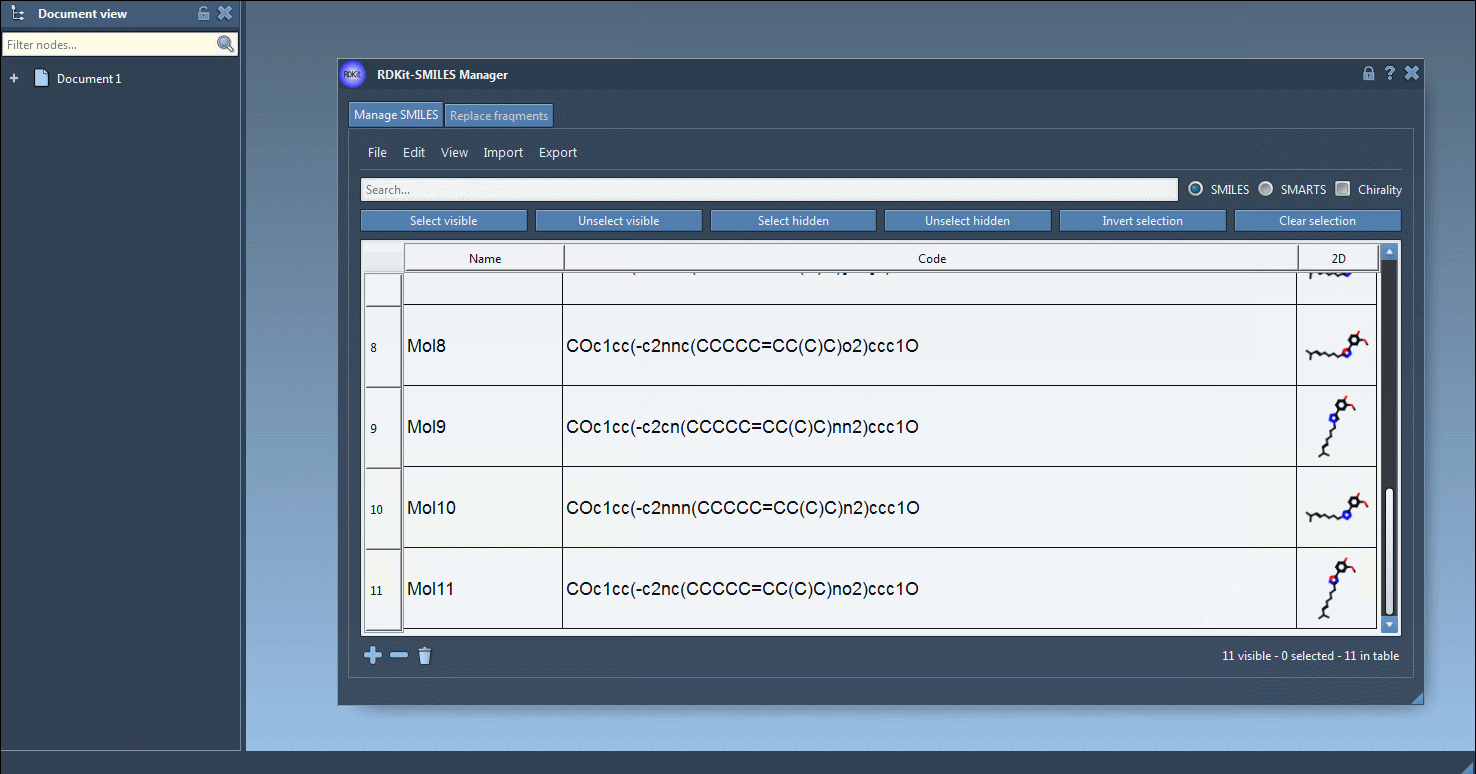
- Validate and Visualize 2D
As SMILES data are loaded, 2D depictions are generated. Errors are flagged by a placeholder image, so you can easily spot problematic entries. Double-click an image to view a larger version or right-click for more options.

- Select and Generate 3D Structures
Select SMILES rows from the table and choose Export > Selected SMILES string to Document. This creates a 3D version for each selected molecule directly in your SAMSON workspace.
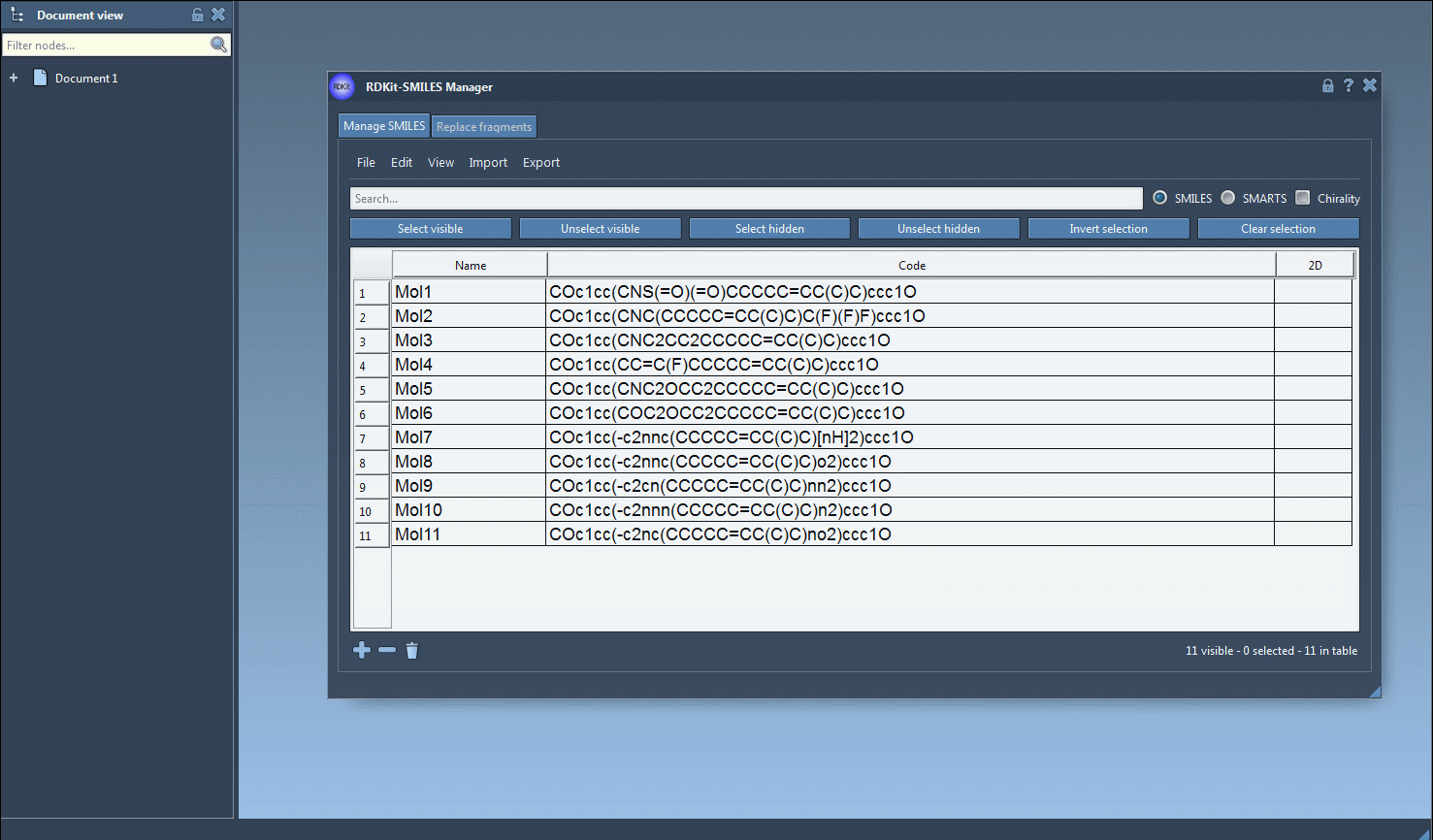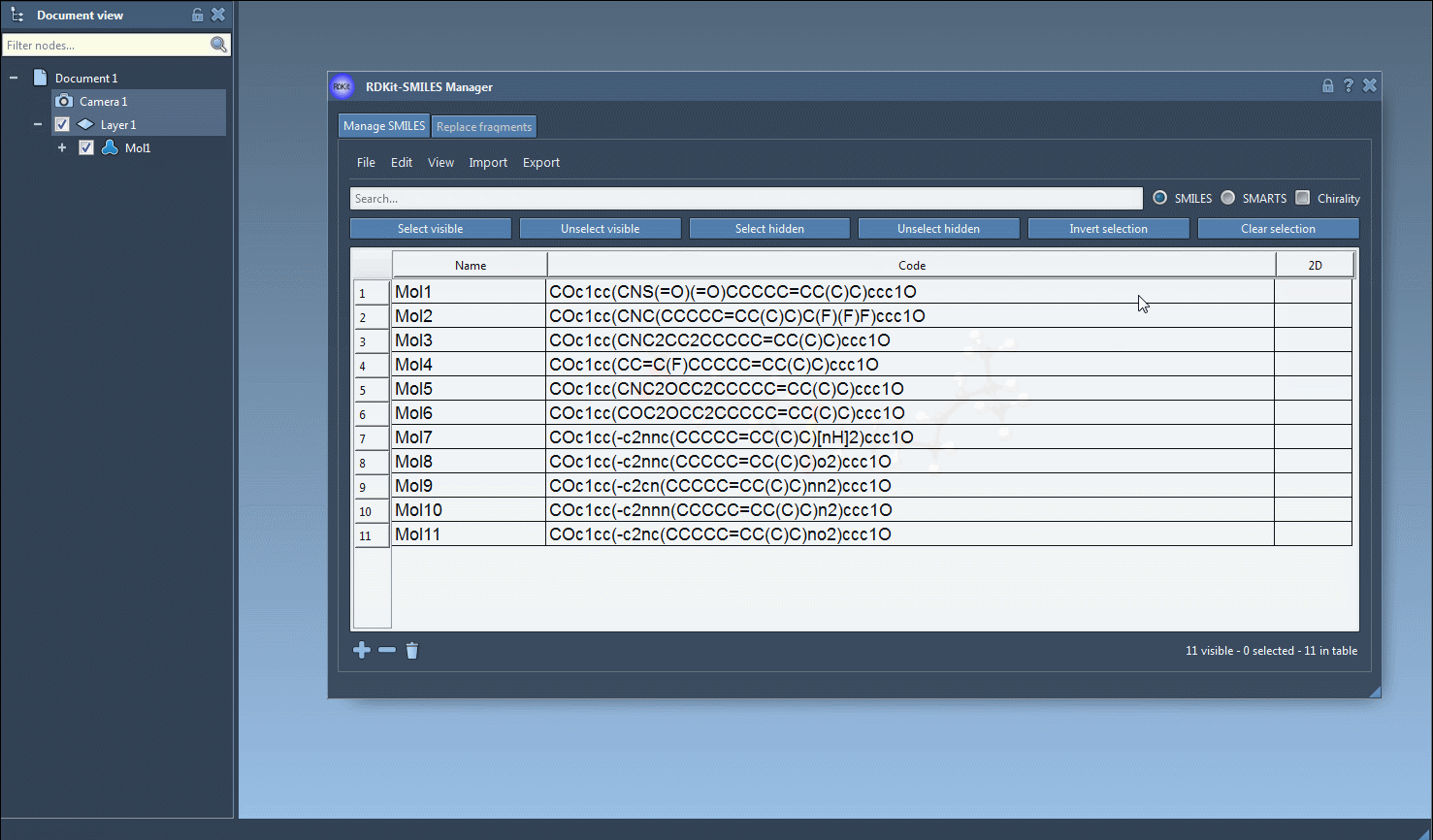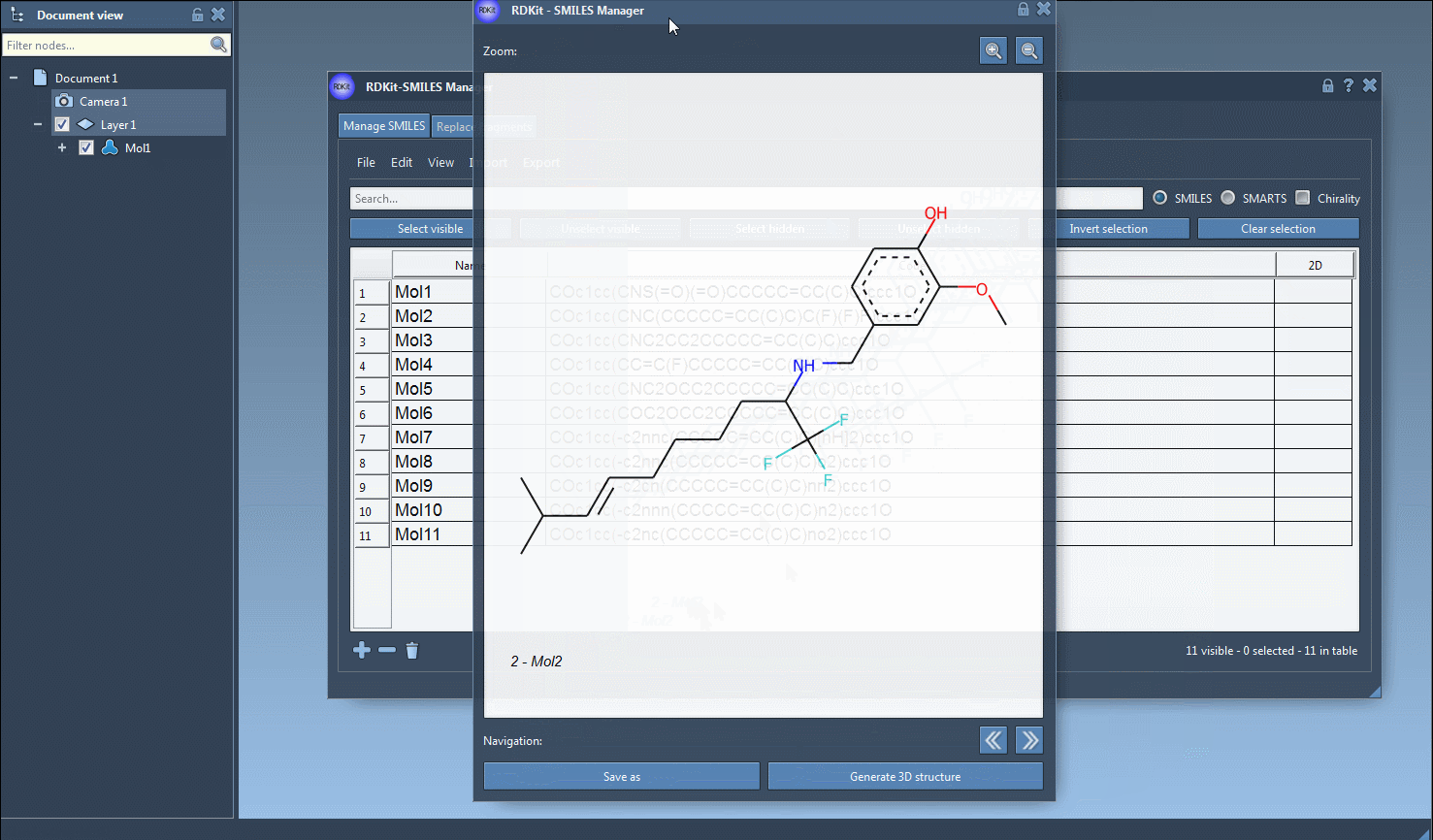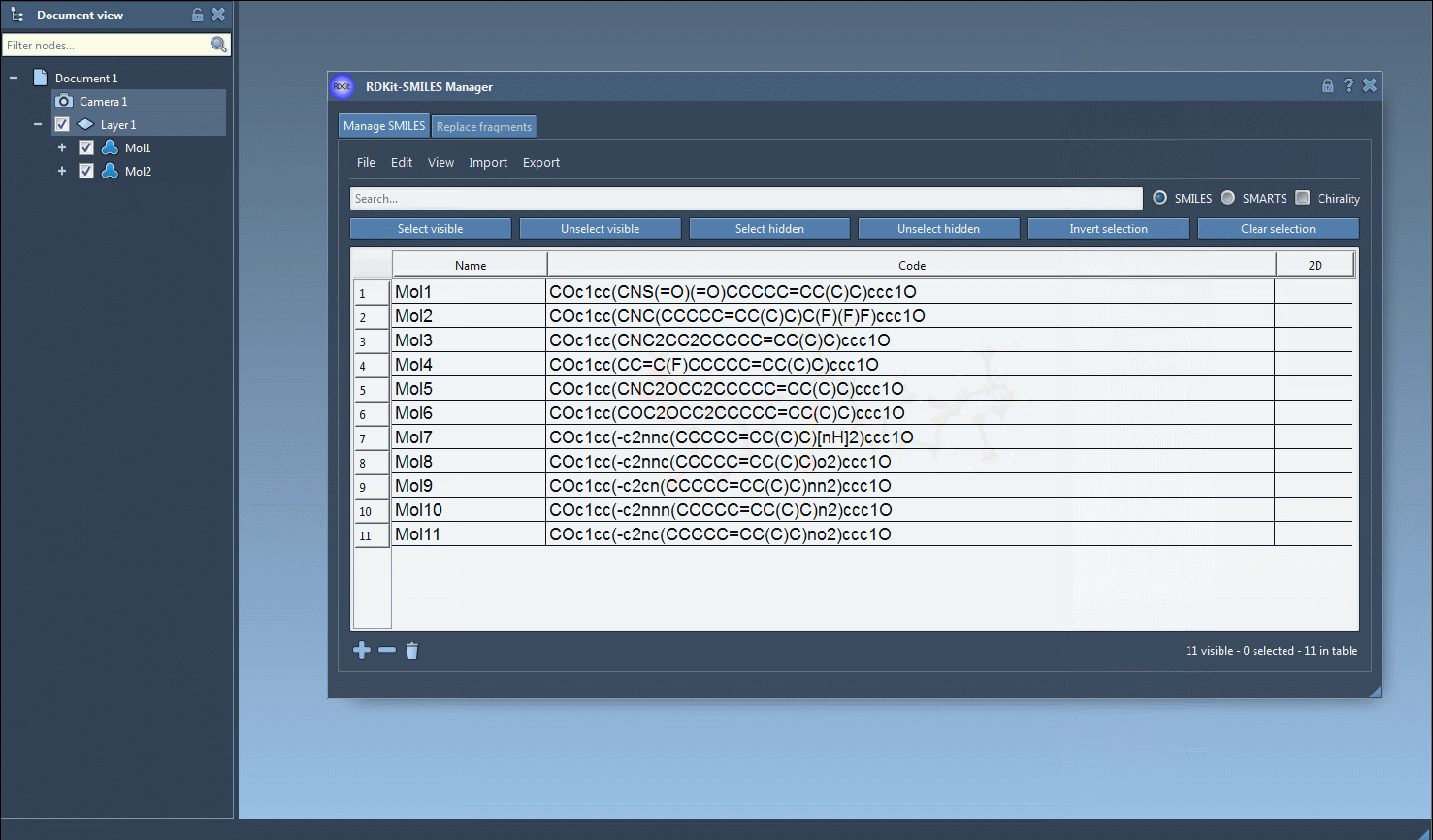
- Single-molecule 3D views
For focused analysis, use the context menu (right-click on a 2D image) or the larger image view window to generate a 3D structure for an individual entry.
Useful Tips
- The 3D structures generated are easily editable and can be exported or used in downstream calculations.
- If you need to generate multiple entries and inspect or tweak them visually, open multiple 3D view panels for better comparison.
- No 2D sketching or external converters needed—the pipeline is entirely visual and reproducible inside SAMSON.
When This Helps Most
If you’re regularly:
- Testing large libraries of candidate molecules
- Converting SMILES for structure-based modeling
- Preparing input for quantum or empirical calculations
then this feature removes friction, avoids format mismatch problems, and gets you directly to what matters: molecules you can work with.
To learn more about this feature and others, visit the full SMILES Manager tutorial.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





