Generating biologically realistic environments for membrane proteins is often a challenge in molecular modeling. One frequent task is preparing a lipid bilayer around a protein to simulate its behavior in a membrane-like system. Traditionally, this requires time-consuming scripting or the use of multiple tools. Fortunately, the Molecular Box Builder in SAMSON simplifies this workflow without sacrificing control or detail.
This post provides a practical guide to assembling a single or double-layer lipid environment around a protein directly within SAMSON. The process is highly visual, does not require programming, and allows for precise alignment and control at every step.
Step 1: Prepare and Align the Protein
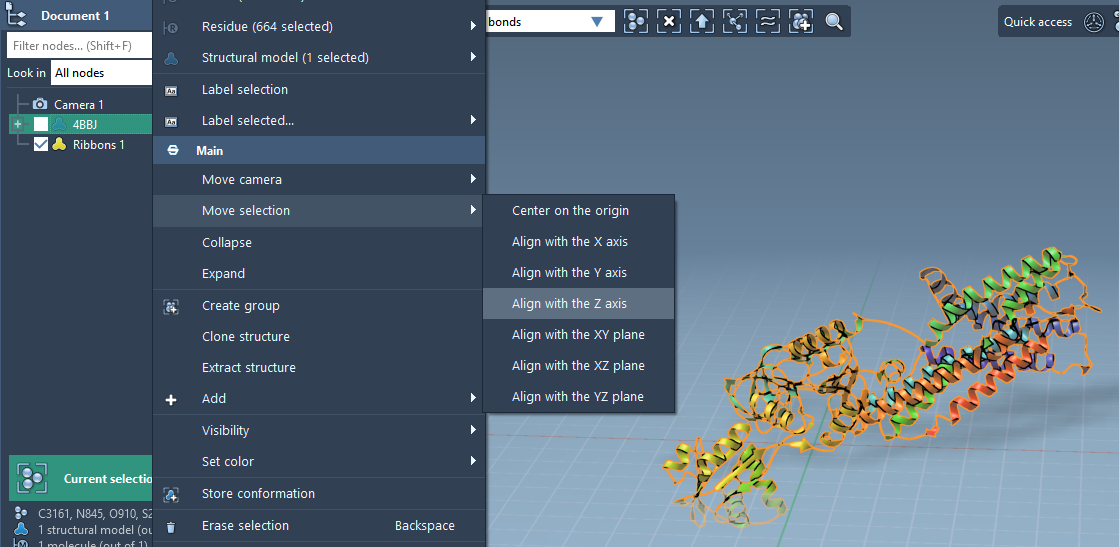
Before adding lipids, it’s essential to orient the protein appropriately. In SAMSON:
- Right-click your protein in the Document view.
- Select Move selection > Align with Z axis to mimic the orientation typical of membrane insertion.
- Then choose Move selection > Center on the origin to have the protein well-positioned for box creation.
Step 2: Select and Align the Lipid Molecule
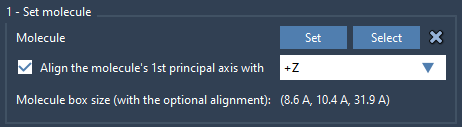
Next, you’ll need a lipid structure—any standard lipid like POPC will do. Import it and:
- Select it in the Viewport or Document View.
- Click Set in the Molecular Box Builder app to use it for populating the box.
- Align the lipid’s axis with
+Zso its orientation fits the membrane geometry.
Step 3: Define the Lipid Layer Box
With both protein and lipid in position, configure the box that will contain the lipids:
- Center the box around the protein coordinates.
- Set its height (Z-dimension) to accommodate a single lipid layer.
- Adjust X and Y dimensions to define the membrane patch size.
- Optionally customize the margin between molecules to control packing density.
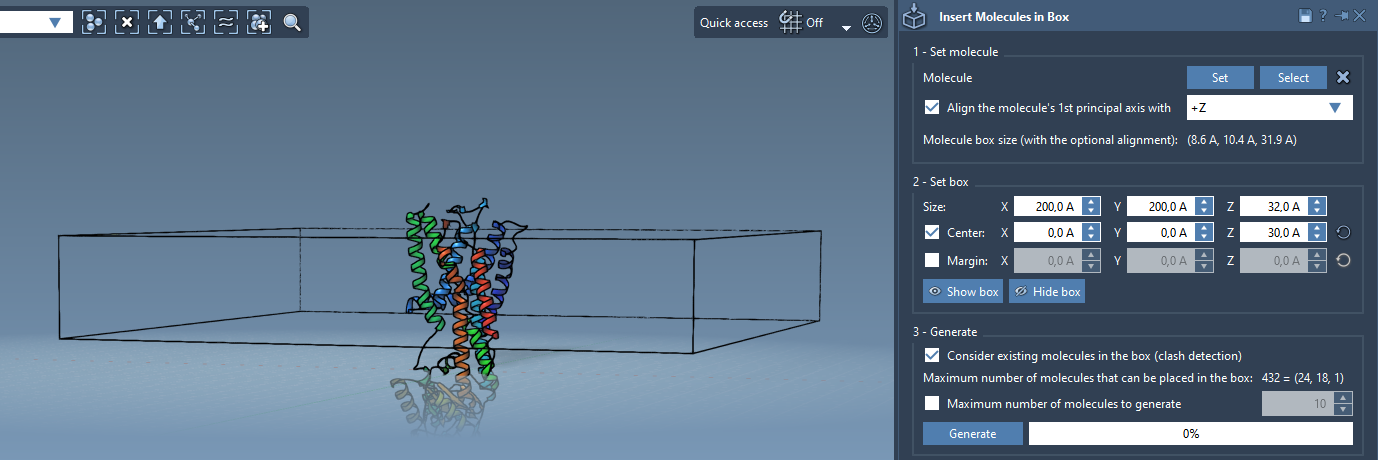
Step 4: Generate the Lipid Layer
To ensure lipids are only added around the protein and not overlapping with it, enable Consider existing molecules in the box. Then click Generate. Within a few seconds, the space is filled with non-overlapping lipids placed around the membrane-facing region of your protein.

Optional: Build a Full Bilayer
Building a bilayer involves repeating the lipid insertion step with reversed alignment:
- Add the first monolayer with
+Zlipid orientation. - Offset the box center in the Z-direction.
- Re-align lipids to
-Zand generate the opposite-facing layer.
This approach allows users to control spacing and composition for each layer independently—helpful when modeling asymmetric membranes.
Conclusion
Creating realistic membrane environments no longer needs to involve multiple tools or manual scripting. With the Molecular Box Builder in SAMSON, a visual and interactive workflow allows you to align, build, and populate molecular systems with precision—from solvent boxes to complex bilayers.
Want to go further? After building the membrane, you can equilibrate and simulate your system using the GROMACS Wizard extension.
To learn more and follow the complete tutorial, visit the original SAMSON Molecular Box Builder Documentation.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





