When designing polymers for simulations or material property prediction, it’s often essential to define precise patterns of monomers: not just their identity but the way they connect. This is particularly true if you’re modeling complex materials, conjugated compounds, or drug-polymer conjugates. In SAMSON’s Polymer Builder, defining and connecting monomer sequences is a flexible process — and understanding this step can save you hours downstream.
Let’s explore how to register and connect custom monomer sequences, especially when you need fine control over bond types like single, double, or triple bonds. If you’re new to Polymer Builder, learning this part alone will help you design a wide range of molecular architectures.
Defining Monomer Sequences
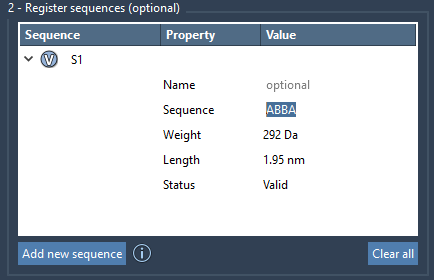
Once you’ve registered individual monomers (fragment units identified by letters like A, B, etc.), you can compose them into sequences using the Register Sequences panel. Each entry is a specific arrangement of monomer IDs, such as:
|
1 |
ABBA |
You can also provide sequences with custom names and view which monomers each sequence includes by clicking their V (View) buttons. The system automatically flags incorrect sequences. For valid ones, predicted values like total molecular weight and length are shown.
Controlling Bond Types Between Monomers
One often overlooked feature is how Polymer Builder lets you control the bond type between fragments. By default, monomers are connected with single bonds. But you can easily specify:
=for a double bond#for a triple bond
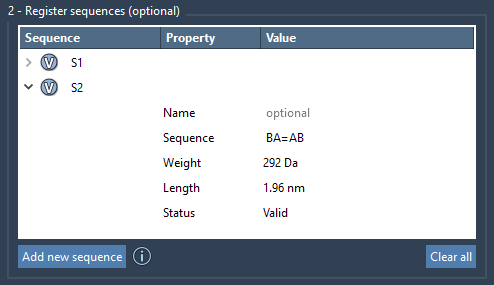
For instance, to create a structure where A and B connect via a double bond and B and C via a triple bond, enter:
|
1 |
A=B#C |
This enabling step unlocks the modeling of alternating single-double structures — key for conjugated polymers — or even rigid linear backbones needed in high-performance materials. Here’s an example view of registered sequences:
Combine, Repeat, and Preview
You can build the final polymer by combining monomer sequences and applying repeat counts. For instance:
|
1 |
2*S1 + AB=BA |
This results in two repeats of sequence S1 joined to sequence AB and BA connected via a double bond. Polymer Builder displays an estimated molecular weight and length before polymer creation:
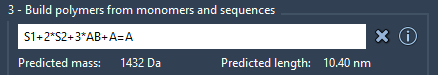
This preview is particularly helpful for quickly assessing whether the design meets your property or simulation constraints without running a full build every time.
Final Notes
By learning how to register monomer sequences and fine-tune their connections, you unlock a powerful combinatorial tool in your molecular design workflow. Whether building alternating copolymers, segment-blocked structures, or drug conjugates, this approach improves both control and reproducibility in your models.
To explore the full capabilities of Polymer Builder, including generating and minimizing full polymer structures, visit the full documentation: https://documentation.samson-connect.net/tutorials/polymer-builder/polymer-builder/
SAMSON and all SAMSON Extensions are free for non-commercial use. Get SAMSON at https://www.samson-connect.net.





