Tracking the motion of molecules in a system can be crucial for understanding mechanisms like ligand binding and unbinding, conformational changes, or for generating reaction coordinate files in free energy methods. If you’ve been manually extracting coordinates along a path, or running additional simulations just to get an atom selection over time, there’s an easier way — directly exporting atom trajectories from path data using SAMSON’s Export Along Paths app.
Let’s take a common use case: you’ve used a ligand path prediction tool (e.g., Ligand Path Finder) in SAMSON to identify how your ligand leaves the binding pocket. Now, you want to perform a series of free energy calculations along this path — which means you need atomic positions, frame by frame.
What this solves 🧪
Typically, exporting trajectory data involves rerunning complex molecular dynamics setups, tedious scripting, or coarse approximations. This SAMSON extension gives a direct way to export just what you need, whether it’s all atoms or a specific group, such as a ligand or a binding pocket.
Exporting a Subset of Atoms — Step by Step
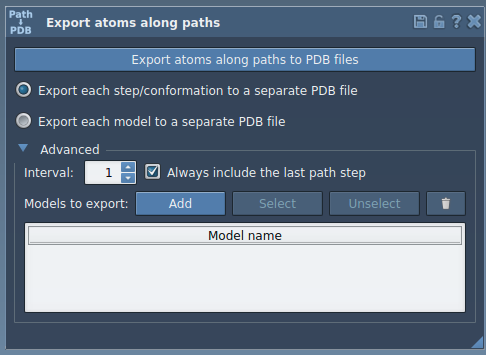
Here’s how to extract the trajectory of a specific molecule (for instance, the ligand TDG) along a computed path.
- Launch the Export Along Paths app from
Home > Apps > All > Export Along Paths. - Expand the Advanced panel in the app interface.
- In the Document View, select the group of atoms you wish to export. For this example, it’s the ligand
TDG.

- Click Add to define this selection as a “model” to export. This creates a named entry in the model table.

You can
- Rename this model by double-clicking its name in the table.
- Add multiple selections (e.g., protein backbone, active site residues).
- Refine or reset selection as needed.
Final Steps
- Select the path(s) you want to export (this may be an unbinding trajectory previously computed).
- Choose whether you want all frames in one PDB file, or each frame in a separate file.
- Click Export atoms along paths to PDB files.
You’ll be asked to choose a destination folder and file prefix. That’s it — you now have a set of PDB files representing how your selected atoms move along the path.
When is this useful?
- Creating reaction coordinate trajectories for umbrella sampling.
- Exporting snapshots for enhanced sampling techniques.
- Getting intermediate 3D states for visualization or presentations.
- Tracking motion of a ligand only, avoiding bulk export of irrelevant atoms.
To learn more about this feature in context and explore further workflow options, visit the original documentation:
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





