When studying molecular mechanisms like ligand unbinding, conformational transitions, or sampling along reaction coordinates, one common challenge arises: how to extract atomic trajectories for further analysis or input to other software packages.
For researchers using SAMSON, the Export Along Paths extension offers a practical solution. This post walks you through how to export atomic trajectories, focusing on a scenario many molecular modelers face—extracting the coordinates of a ligand as it exits a protein, following a defined path.
Why export trajectories?
Whether you are preparing input for free energy calculations, need intermediate conformations for visualization, or are scripting enhanced sampling runs, exporting atomic coordinates is often an essential step. However, most molecular software lacks an easy mechanism for exporting atoms along complex paths—this is where SAMSON steps in.
Case Study: Ligand Unbinding Path
In this example, we begin with a sample document containing Lactose permease (PDB: 1PV7) and its ligand Thiodigalactosid (TDG). The ligand unbinding path has already been computed using the Ligand Path Finder extension.
Here’s how to export just the TDG ligand’s atomic coordinates along this path:
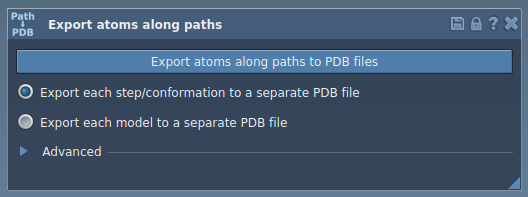
Step 1: Open the Export Along Paths App
Go to Home > Apps > All > Export Along Paths or press Shift+E. The app allows you to manage export modes and choose which atoms to include.
Step 2: Select the Ligand
In the Document view, select the ligand named TDG. You can right-click it or use the selection tools.

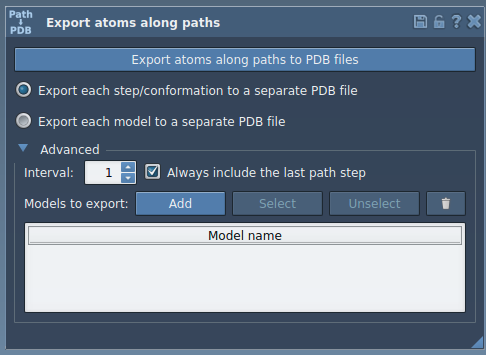
Step 3: Add it to the Export List
With the ligand selected, open the Advanced section of the app. Click Add to define the TDG molecule as a model to export. This allows you to specify a subset of atoms instead of the entire system.
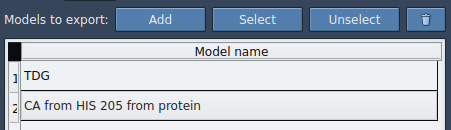
A new entry appears in your model list. Here, you can rename it, highlight included atoms, and reset your selection if needed.
Step 4: Choose Export Format
- All frames in one PDB file
- One PDB file per frame
Choose the appropriate format depending on your intended use—e.g., scripting analysis tools or generating simulation intermediates.
Step 5: Select the Path and Export
Select one or more unbinding paths from the Document view. Then click Export atoms along paths to PDB files. You’ll be prompted to select a folder and file prefix.
That’s it. You now have coordinate files of your selected atom subset along a predefined path—ideal for integration with other tools in your workflow.
Tip: Add Multiple Atom Sets
You’re not limited to one molecule. You can add multiple sets of atoms (e.g., ligand + surrounding residues) and export their joint trajectories along the same path.
Who is this for?
This process is especially helpful for users performing:
- Reaction coordinate generation
- Free energy profiling
- Trajectory analysis and visualization
To learn more and explore advanced features, visit the full documentation page: Export Atom Trajectories Along Paths.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





