One recurring challenge for molecular modelers is understanding how large biomolecular systems transition between two experimentally known states. In many cases, only the starting and ending conformations of a molecular structure are available—leaving open the question of how the system evolves between these configurations. This problem is especially relevant when studying essential motions in virology, such as the opening of the SARS-CoV-2 spike protein to bind to the ACE2 receptor.
In this post, we’ll explore how the SAMSON integrative molecular design platform allows you to animate such transitions and generate intermediate conformations using the case of the SARS-CoV-2 spike protein opening motion.
The challenge: connecting open and closed states
Transition modeling can be especially tricky when working with structures that differ in the number of residues. This can prevent the use of simple interpolation techniques and often forces researchers to resort to complex manual modeling workflows.
However, using SAMSON’s ARAP Interpolation Path and Parallel Nudged Elastic Band (P-NEB) modules, you can generate reasonable trajectories even when residue counts differ—as is the case between the spike’s closed (PDB ID: 6VXX) and open (PDB ID: 6VYB) structures.
Workflow overview: how the motion was calculated
- Structure preparation: Modified bond orders in sugar molecules using a custom Python script, then added hydrogens and minimized both structures (6VXX and 6VYB) inside SAMSON.
- Initial interpolation: Generated an interpolated path using the As-Rigid-As-Possible (ARAP) module, going from the open state as the starting conformation to the closed state as the goal conformation.
- Residue mismatch fix: Since the original two structures contain a different number of residues, an intermediate conformation from the ARAP path that shares the same residue set as 6VYB was selected and used in a second ARAP run.
- Path refinement: Finally, post-processed the ARAP path with the P-NEB module to generate a smoother, energetically refined trajectory, which finished in about 15 minutes on a laptop.
Visualizing the motion
The result is a set of intermediate structures showing how the receptor-binding domain of the spike opens to expose the ACE2 binding site. These conformations give researchers a powerful starting point to identify intermediate states, study dynamics, and potentially assess candidate binding interactions for drug or antibody design.
Here are some views of the animated opening of the spike:
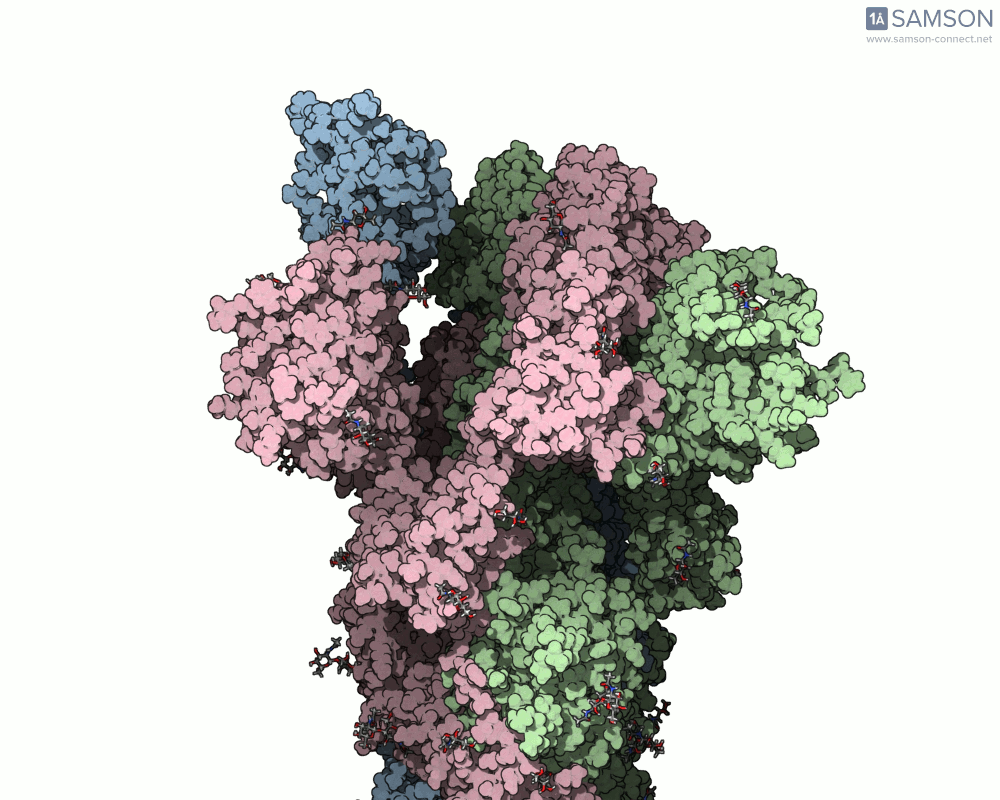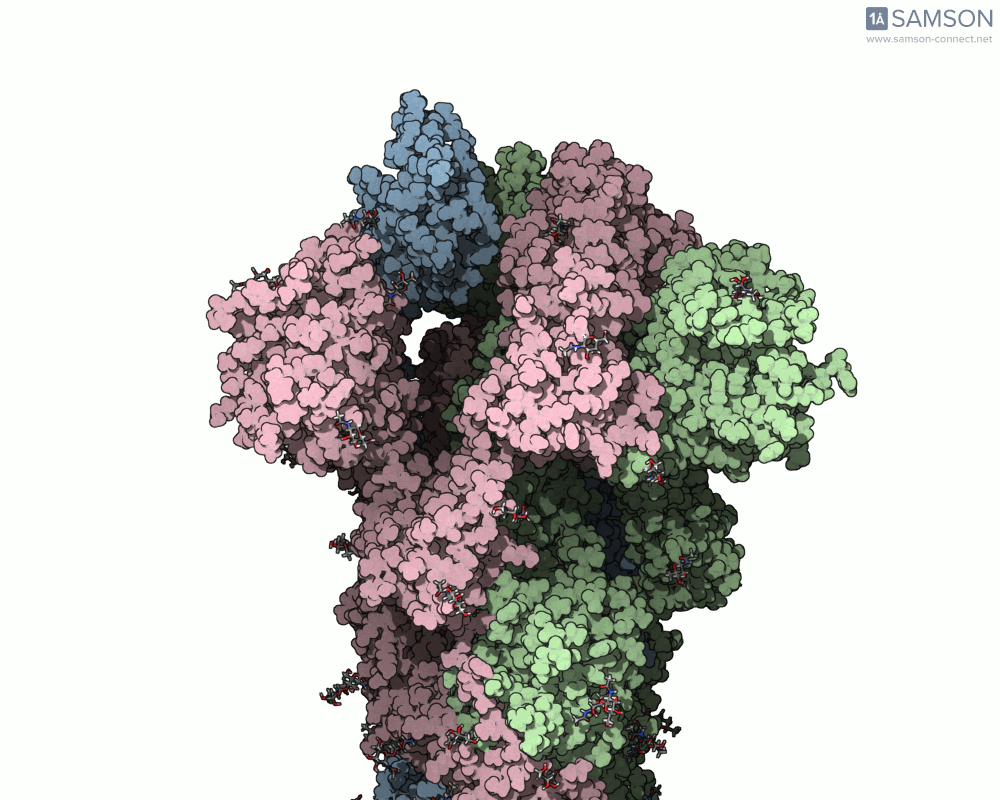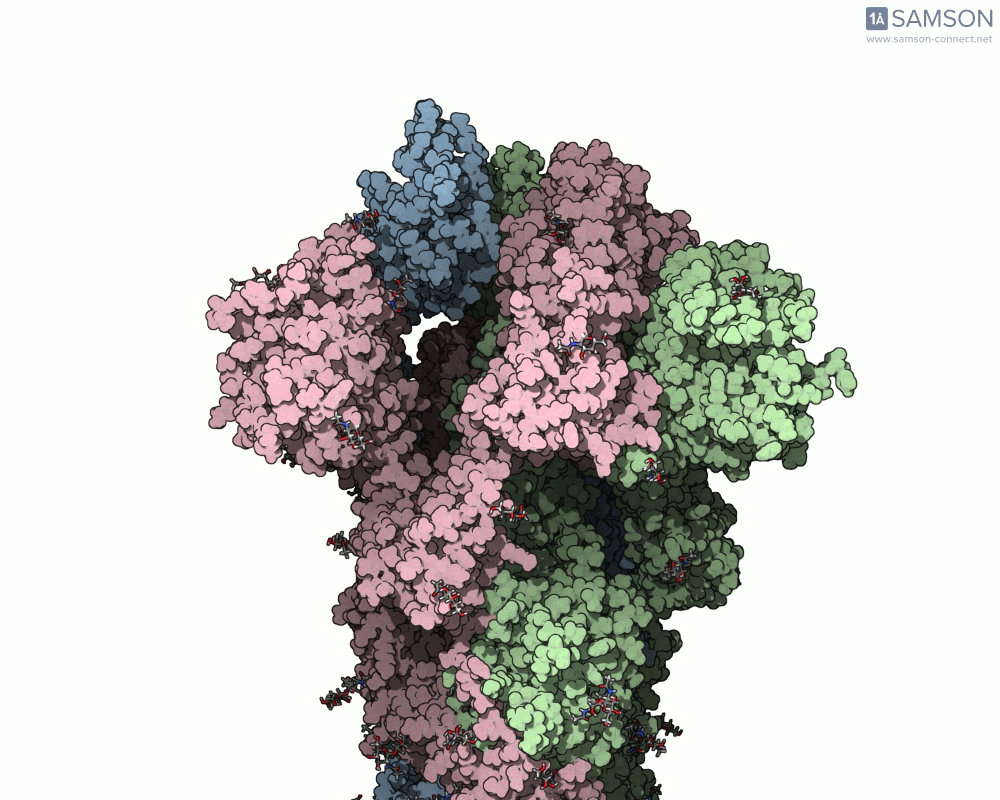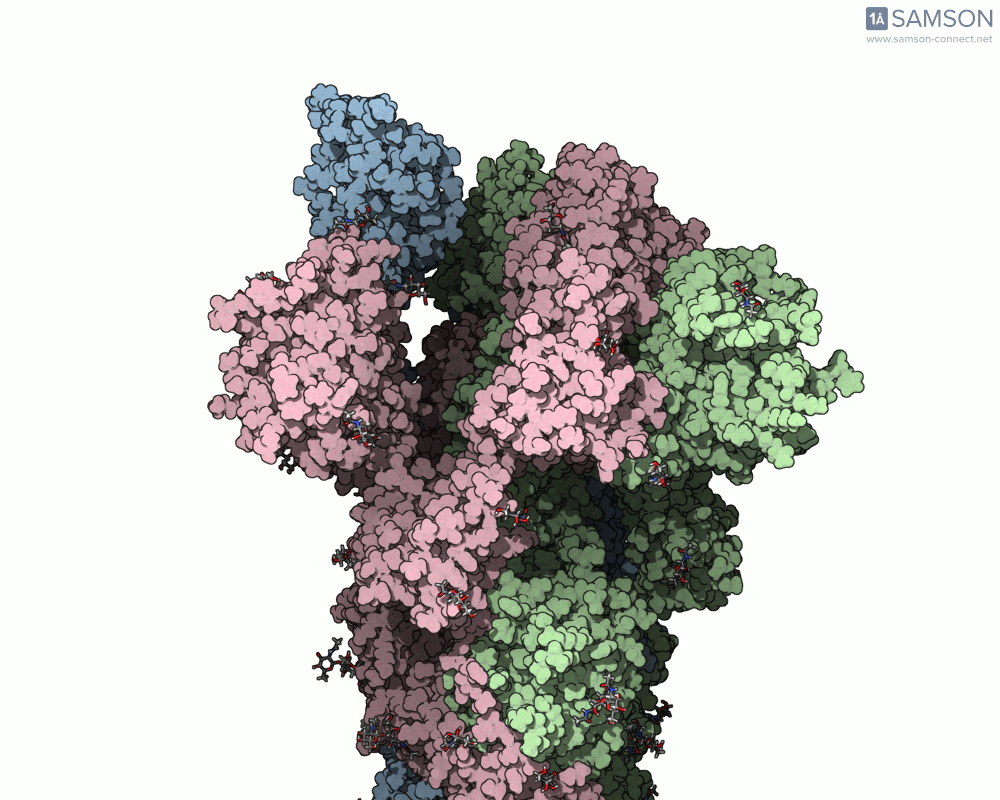
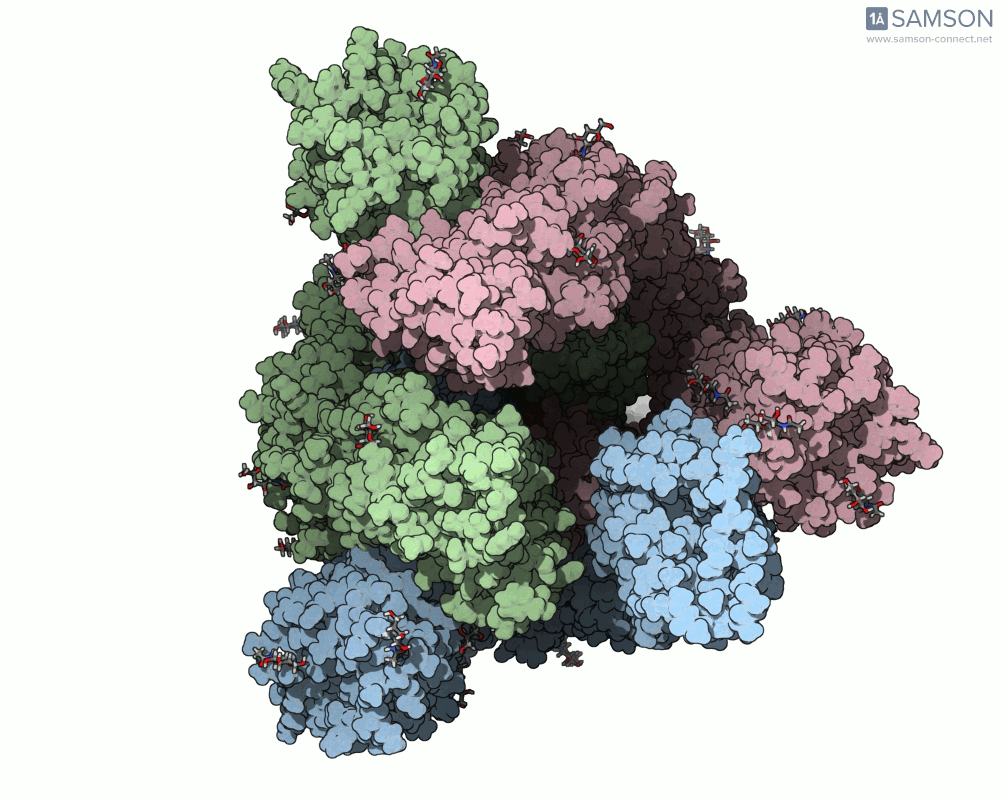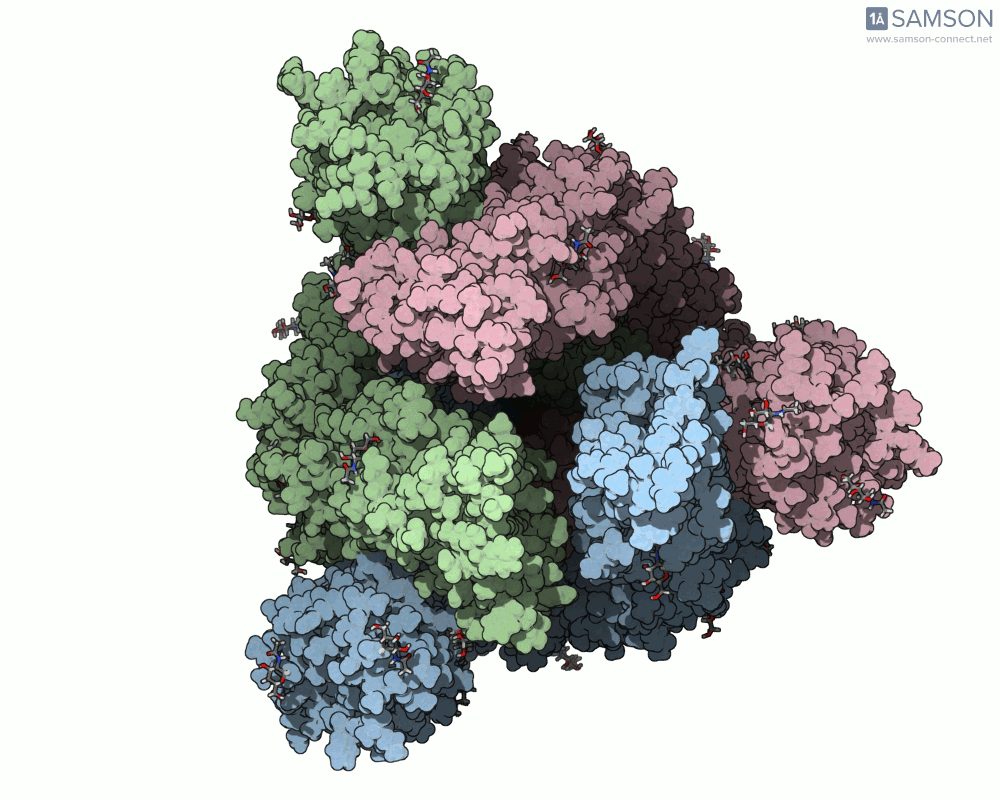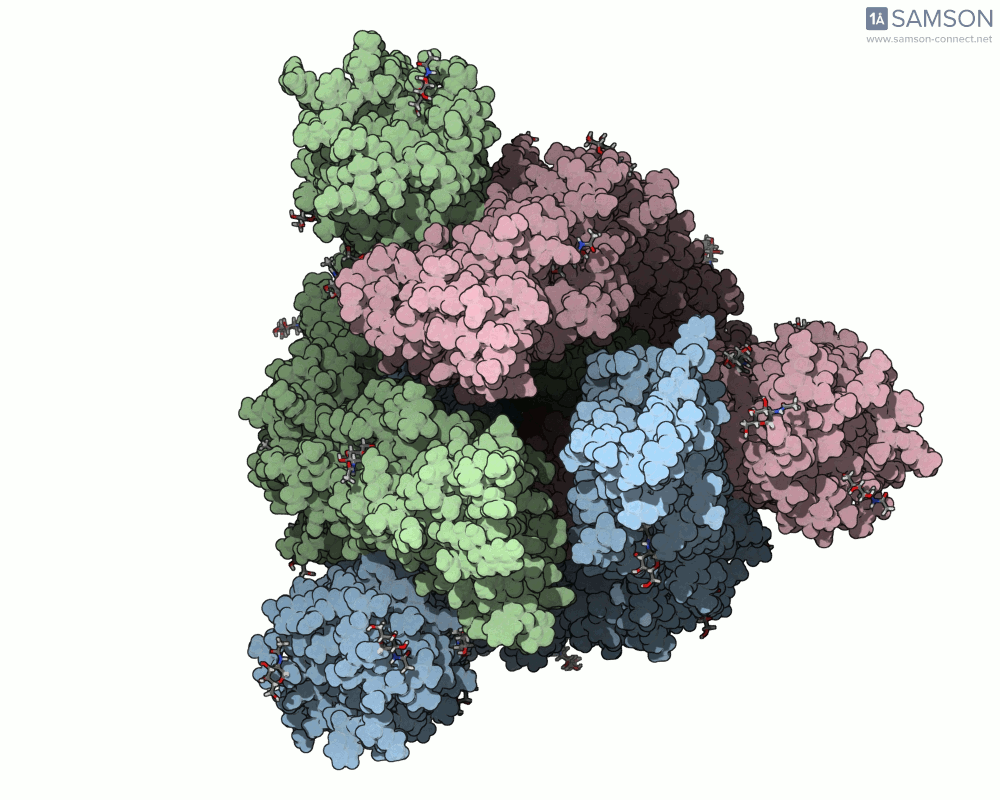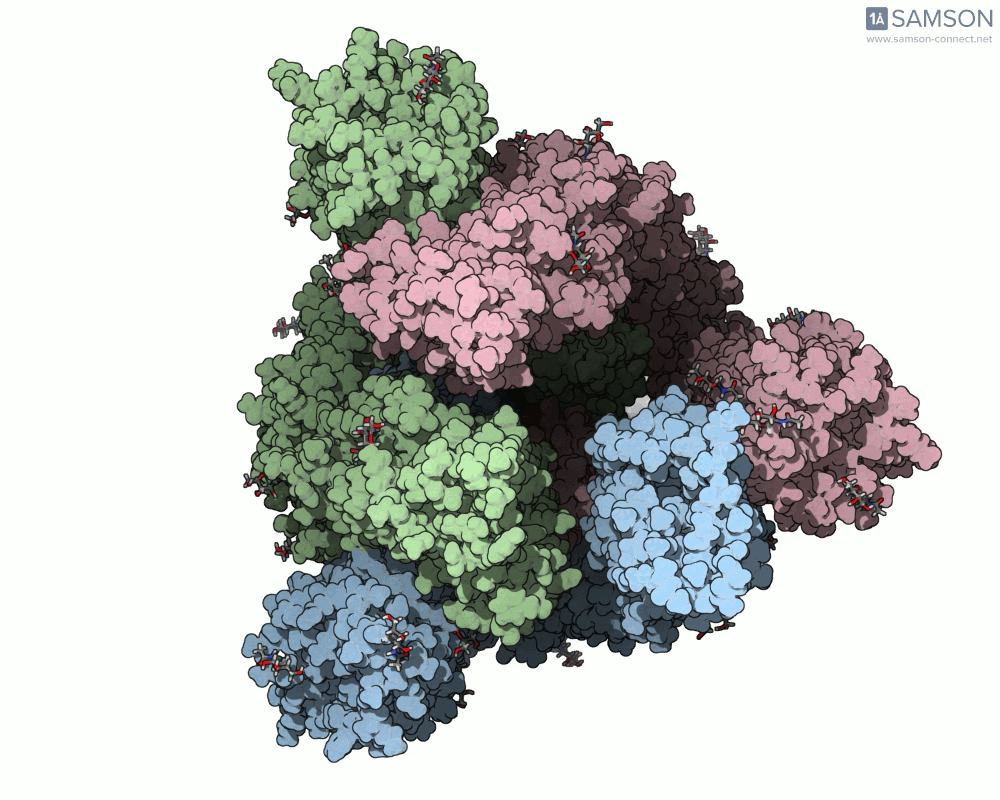
These trajectories illustrate the transition from the closed (down) to open (up) states that enable receptor binding.
Try it yourself
You can download the trajectories as PDB files or in SAMSON’s own .sam format to analyze them directly within SAMSON. These structures can serve as useful approximations or starting points for further refinement, docking experiments, or molecular dynamics simulations—especially for those designing therapeutics targeting intermediate forms of the spike.
To learn more about the process and access the Python scripts and downloadable files, visit the full documentation page here.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





