Adding lipid bilayers around membrane proteins is often a crucial step in setting up biologically relevant molecular dynamics simulations. But manually constructing these bilayers can be a tedious and error-prone task, especially when trying to avoid overlaps or ensure correct orientation. Fortunately, the Molecular Box Builder extension in SAMSON offers a practical way to generate lipid layers directly around your protein structures with just a few steps.
This short guide outlines how to create a lipid monolayer or bilayer around a membrane protein using Molecular Box Builder.
Preparing the Protein
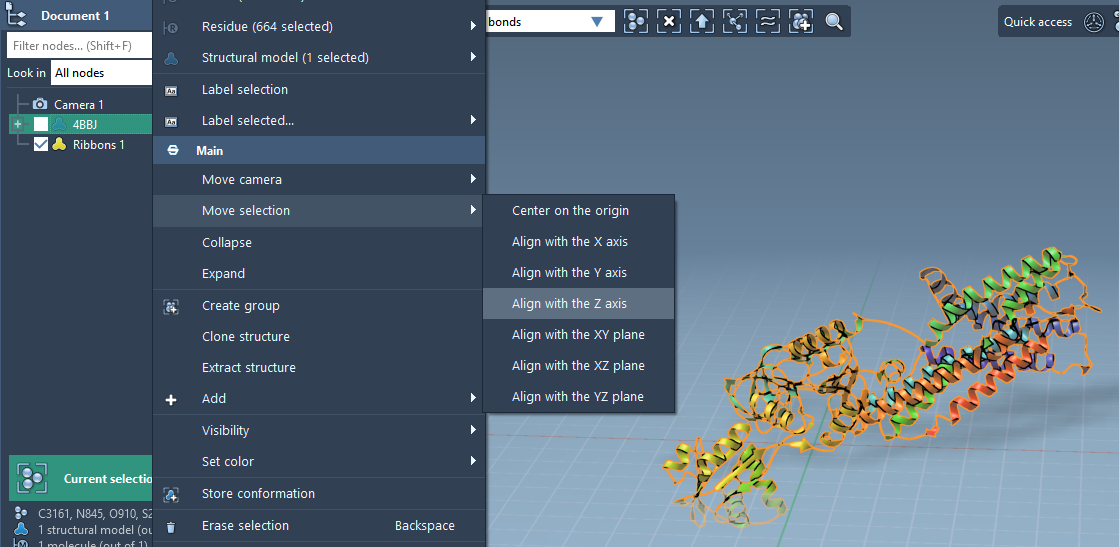
Start by importing or loading your protein structure into SAMSON. To ensure the lipid layer forms correctly around the protein, the structure needs to be aligned along the z-axis and centered around the origin. Here’s how:
- In the Document view, right-click your protein.
- Choose Move selection > Align with Z axis.
- Then, select Move selection > Center on the origin.
Loading and Orienting the Lipid Molecule
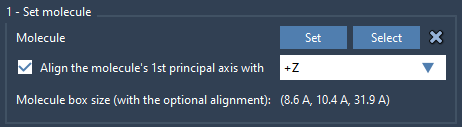
Now import or load a lipid molecule you’d like to use. Select the molecule, then inside the Molecular Box Builder app click Set to designate it as the molecule to insert. To mimic the correct membrane orientation, align its principal axis to the +Z axis using the orientation settings in the same app.
Creating the Lipid Layer Box
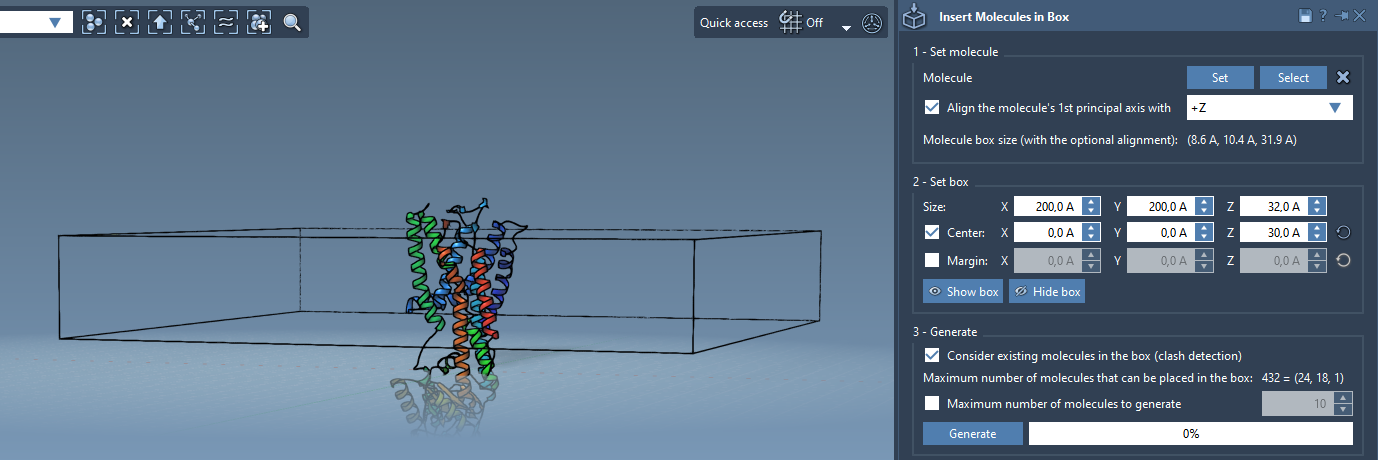
Next, define the box that will contain the inserted lipids. It should surround the protein and be appropriate in thickness for a lipid monolayer. In the app’s box configuration section:
- Choose to center the box around the protein (use the Center option and coordinates accordingly).
- Add margins between the inserted molecules if needed to avoid steric clashes.
Generating the Monolayer
Before generating, make sure to enable the option Consider existing molecules in the box—this ensures the software respects the space already occupied by the protein.
- Click Generate.
You should now see a lipid monolayer surrounding your membrane protein.

Creating a Lipid Bilayer (Optional)
To build a full bilayer:
- First, generate the upper leaflet using
+Zalignment. - Then shift the box center downward in the
Zdirection. - Regenerate the bottom leaflet using the same lipid molecule with
-Zalignment.
This creates a complete bilayer encapsulating the protein, ready for solvating and ion placement.
Final Steps
Once your bilayer system is ready, you can proceed with solvation, neutralization, and molecular dynamics preparation using tools such as the GROMACS Wizard.
Building biologically accurate membrane environments becomes easier with this workflow—and more importantly, reproducible.
To explore the full tutorial and features of Molecular Box Builder, visit the official documentation page: https://documentation.samson-connect.net/tutorials/molecular-box-builder/molecular-box-builder/
SAMSON and all SAMSON Extensions are free for non-commercial use. You can get SAMSON at https://www.samson-connect.net.





