When working with protein conformational changes, it’s often necessary not only to generate realistic transitions between structures, but also to save and share these transitions for visualization, analysis, or subsequent simulation. If you’re using SAMSON’s ARAP Interpolator to create smooth structural transitions, one of the most practical aspects comes at the end of the workflow: exporting your results.
Whether you’re preparing figures for a paper, setting up an umbrella sampling simulation, or simply sharing your results with a colleague, SAMSON provides two clean export options directly within the ARAP Interpolation App. Here’s what you need to know.
Understanding the Path Export Options
After generating your transition path between two protein conformations using ARAP interpolation, scroll to the bottom section of the app interface, where you’ll find two export options:
- Export path: This saves the entire transition trajectory as a path object within your current SAMSON document. It’s a good choice if you want to continue analyzing or modifying the results in SAMSON.
- Export PDB: This writes the interpolated conformations as PDB files. You’ll get one file containing all conformations as sequential models, and individual PDBs for each conformation. This is especially useful for external tools like GROMACS, visualizations in PyMOL, or archiving structures.
What Gets Exported?
Each conformation in the path reflects a snapshot along the interpolation. When you set the number of interpolated conformations (e.g., 20), SAMSON includes the start and end states. So if you’re exporting 20 conformations, that includes the initial and final structures (18 interpolated frames in between).
This makes it easy to create animations or represent structural ensembles step-by-step. The exporter preserves atom information, residue names, and chain identifiers so that your downstream analysis or visualization tools can interpret the files correctly.
Tip: Saving for External Simulations
If your next step involves preparing an umbrella sampling or P-NEB simulation, exporting the path as individual PDBs can drastically streamline the workflow. You can directly input these into the GROMACS Wizard or the P-NEB App for enhanced path optimization or free energy calculations.
Making the Most of Visualizations
Before exporting, take advantage of visualizing the pathlines with the slider and observing edge construction logic. Color-coded edges provide insights into how the structure was interpolated, while toggling visibility in the Document View lets you clean up your working space.
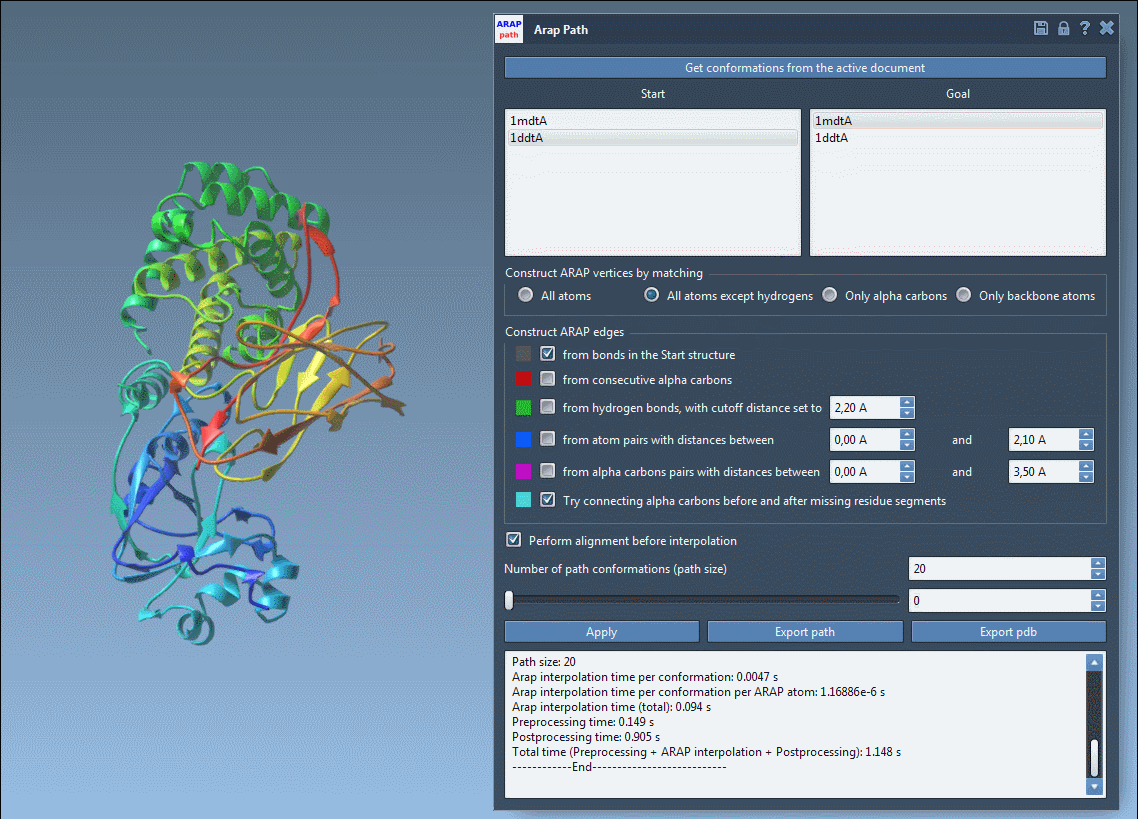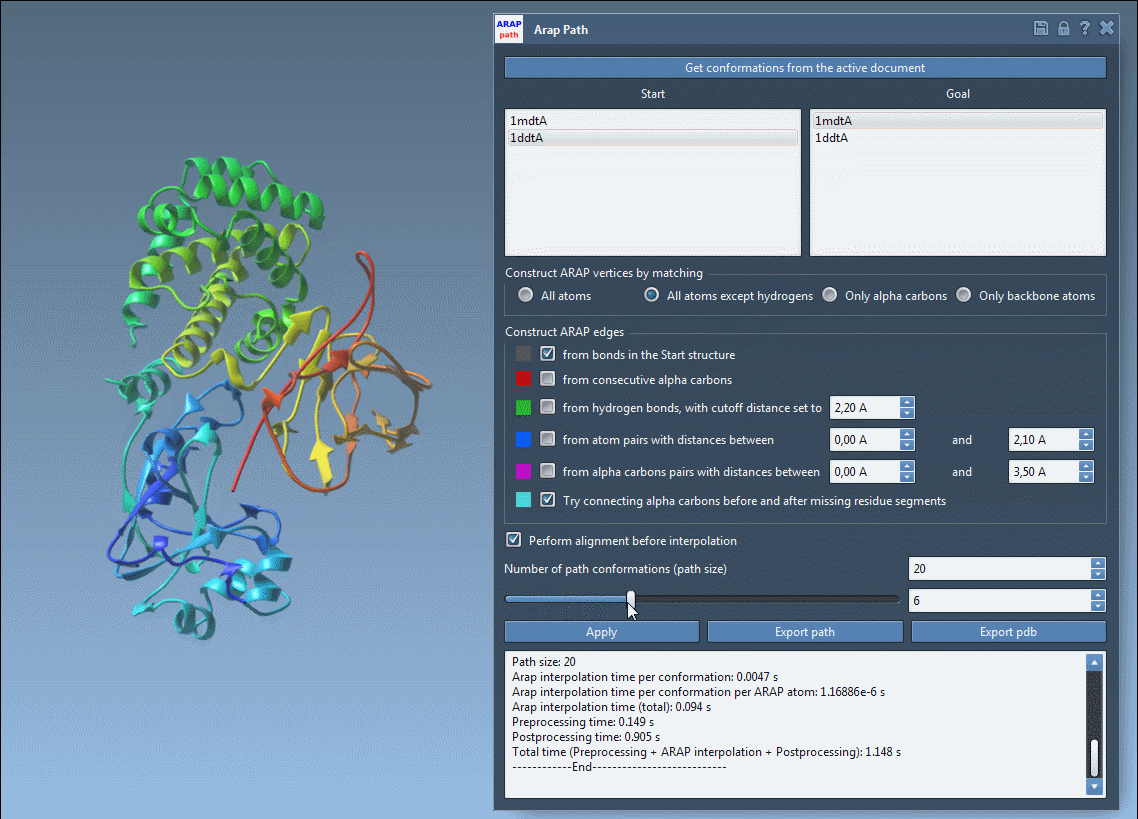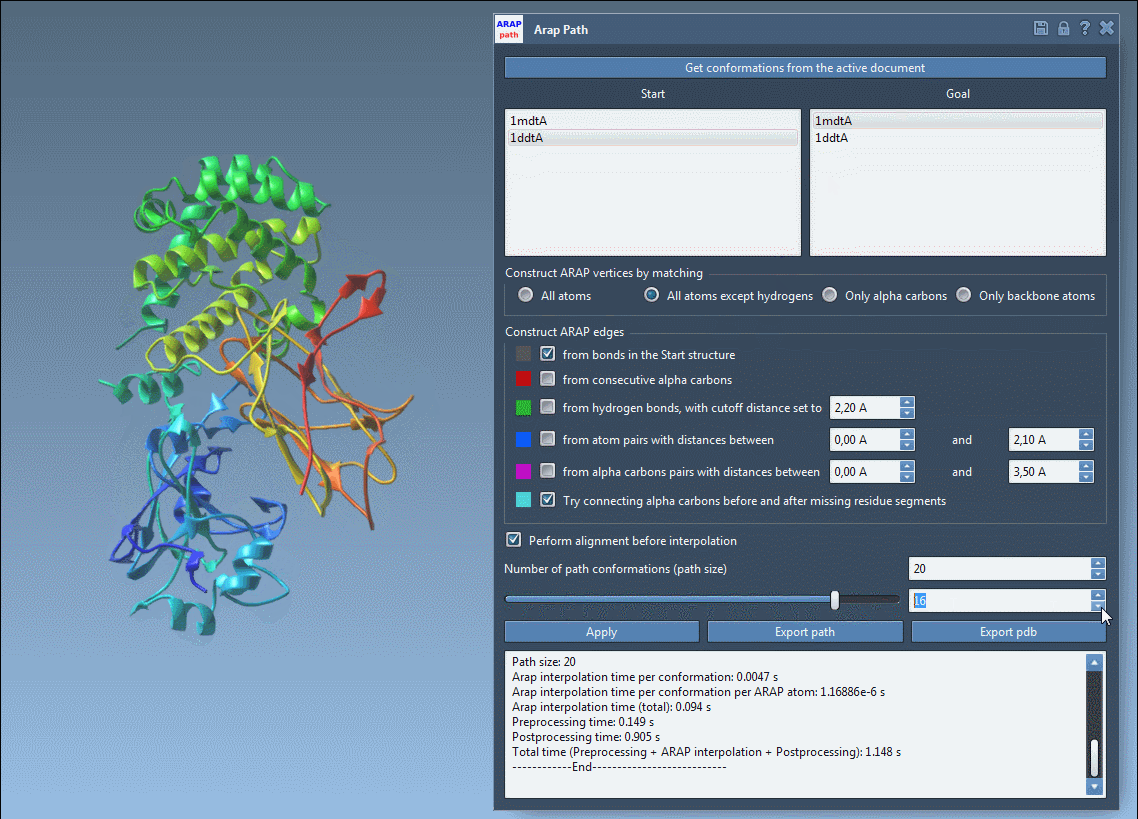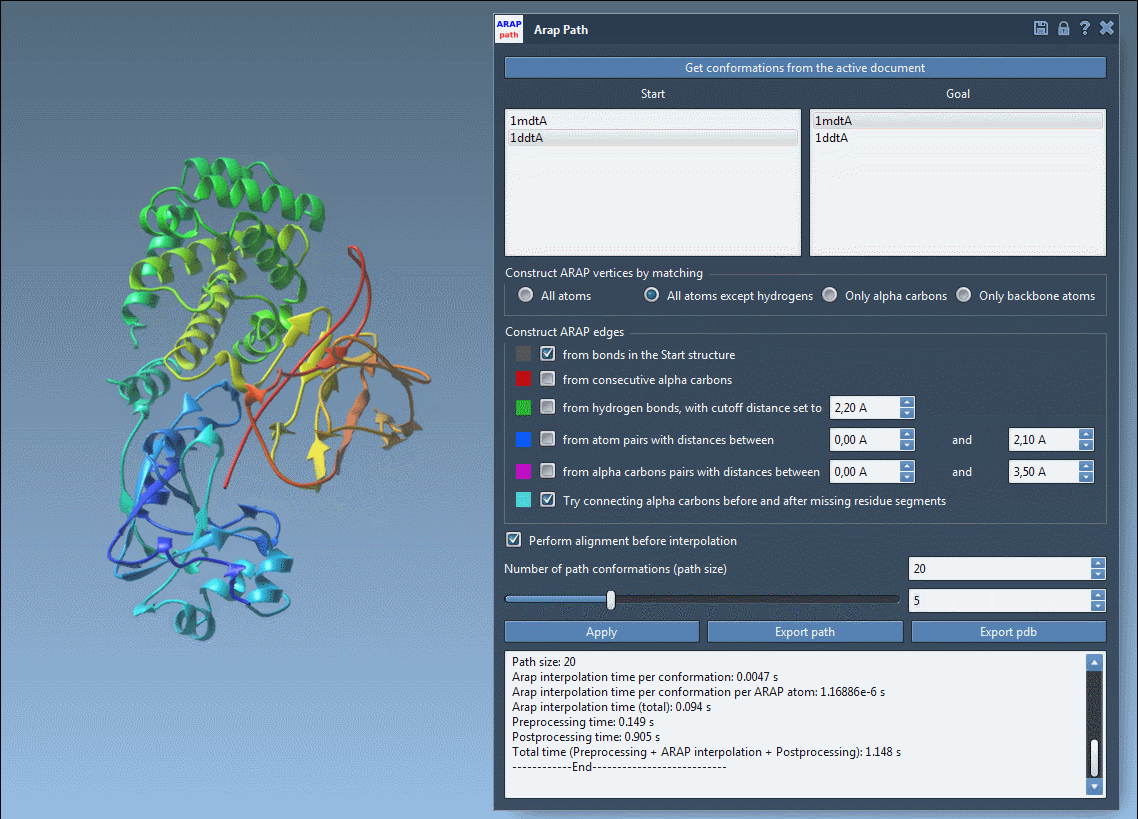
Visual previews help you ensure your transition is clean and biologically meaningful before committing to an export.
Final Thoughts
Although exporting might seem like a final administrative step, it’s one of the most central parts of moving from modeling to communication or simulation. With just a couple of clicks, you can package your work in widely compatible formats and open the door to analysis far beyond the initial interpolation.
To learn how to generate the ARAP interpolation paths and prepare your protein structures, visit the full documentation page: ARAP Interpolation Tutorial.
SAMSON and all SAMSON Extensions are free for non-commercial use. You can download SAMSON at https://www.samson-connect.net.





